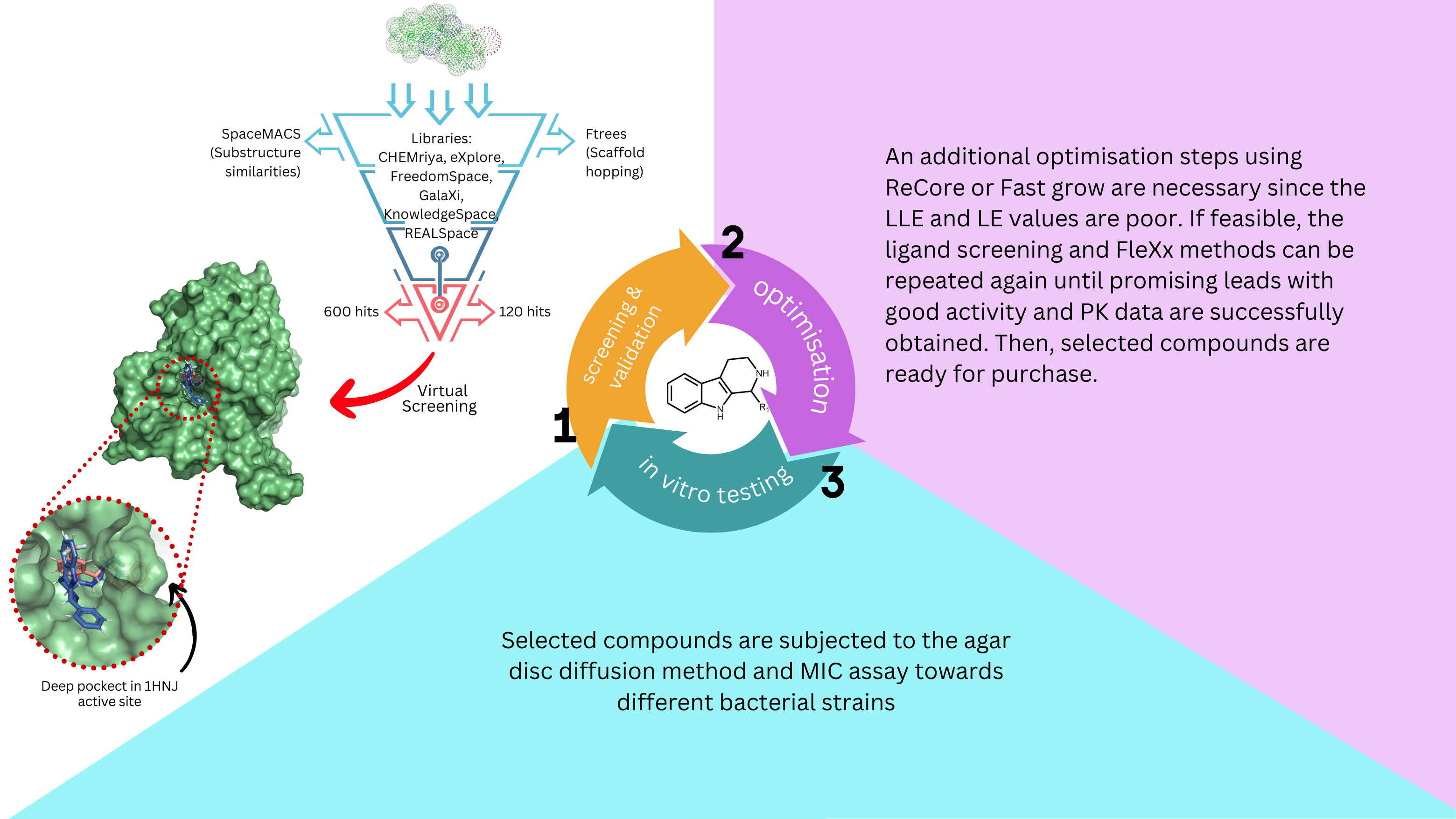
Ligand screening was conducted utilising SpaceMACS and Ftrees to mine substructure similarities and discover new scaffolds for β-carboline, respectively. All libraries were screened, resulting in only 720 hits being identified. Of these, 120 hits are from Ftrees, and the remaining hits come from SpaceMACS. Then, all hits were validated using FlexX towards the FabH protein (1HNJ) to identify which hits can strongly bind to protein active site. The estimated efficiency was ranked from the lowest to the highest and visually inspected to confirm its binding. Most hits showed poor LLE and LE values, but two hits from Ftrees gave improved efficiency ranging from 54-97 nM potencies, surpassing β-carboline. These hits also bind in the deep pocket of the active site, providing additional stability. The top compound from SpaceMACS only gave >1550 nM. Further optimisation are required owing to unsatisfactory LLE values before purchasing compounds.
After 3 months, Nurasyikin has achieved the following milestones:
- Utilising Ftrees is more straightforward and convenient in infiniteSee. Using all available libraries in Ftress, I anticipated a higher number of hits. However, I only received 120 hits probably due to the complexity of my scaffold. For SpaceMACS, the command line is a bit tricky at first, and I need to seek guidance from a Biosolveit team. After several attempts, I managed to obtain the data, but I am struggling to surpass 600 hits due to difficulties in adjusting the limit compound in the provided script. 720 hits obtained from Ftrees and SpaceMACS were then validated using FlexX. Although the hits obtained showed promising compounds, most of them displayed poor LLE and LE values, suggesting the need to address this issue. The visual evaluation of the protein in surface mode is quite challenging, as it’s hard to clearly identify which hits may fill the deep pocket on the active site. Thus, I used Pymol for this purpose.
- Additional optimisation steps using ReCore or Fast Grow are necessary since the LLE and LE values are poor. If feasible, the ligand screening and FleXx methods can be repeated again until promising leads with good activity and PK data are successfully obtained. Then, selected compounds are ready for purchase.
- Selected compounds are subjected to the agar disc diffusion method and MIC assays towards different bacterial strains