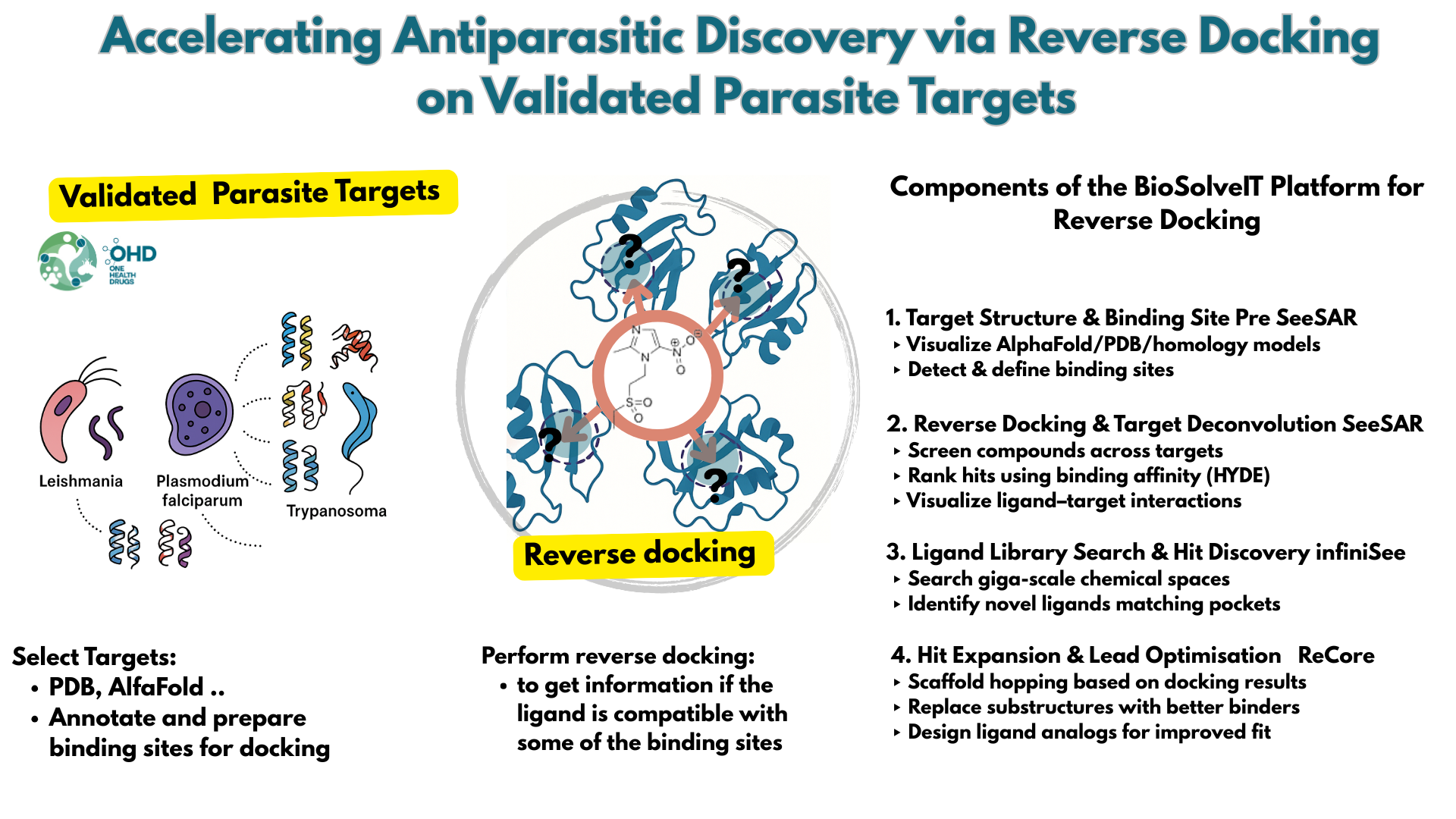
Effortlessly design drug candidates and perform molecular modeling tasks.

Screen ultra-vast Chemical Spaces for relevant compounds based on the needs of the project.

Empower your team with seamless access to high-performance computing.

Access Enamine's largest catalog of accessible and drug-like compounds.
Platforms
SeeSAR
Drug Design Dashboard
infiniSee
Chemical Space Navigator
infiniSee xREAL
xREAL Space Navigator
HPSee
Scalable Workflow Environment
Components
Resources
This website uses cookies so that we can provide the best service possible.
Get more information from our privacy policy.
Accept selected
Accept all