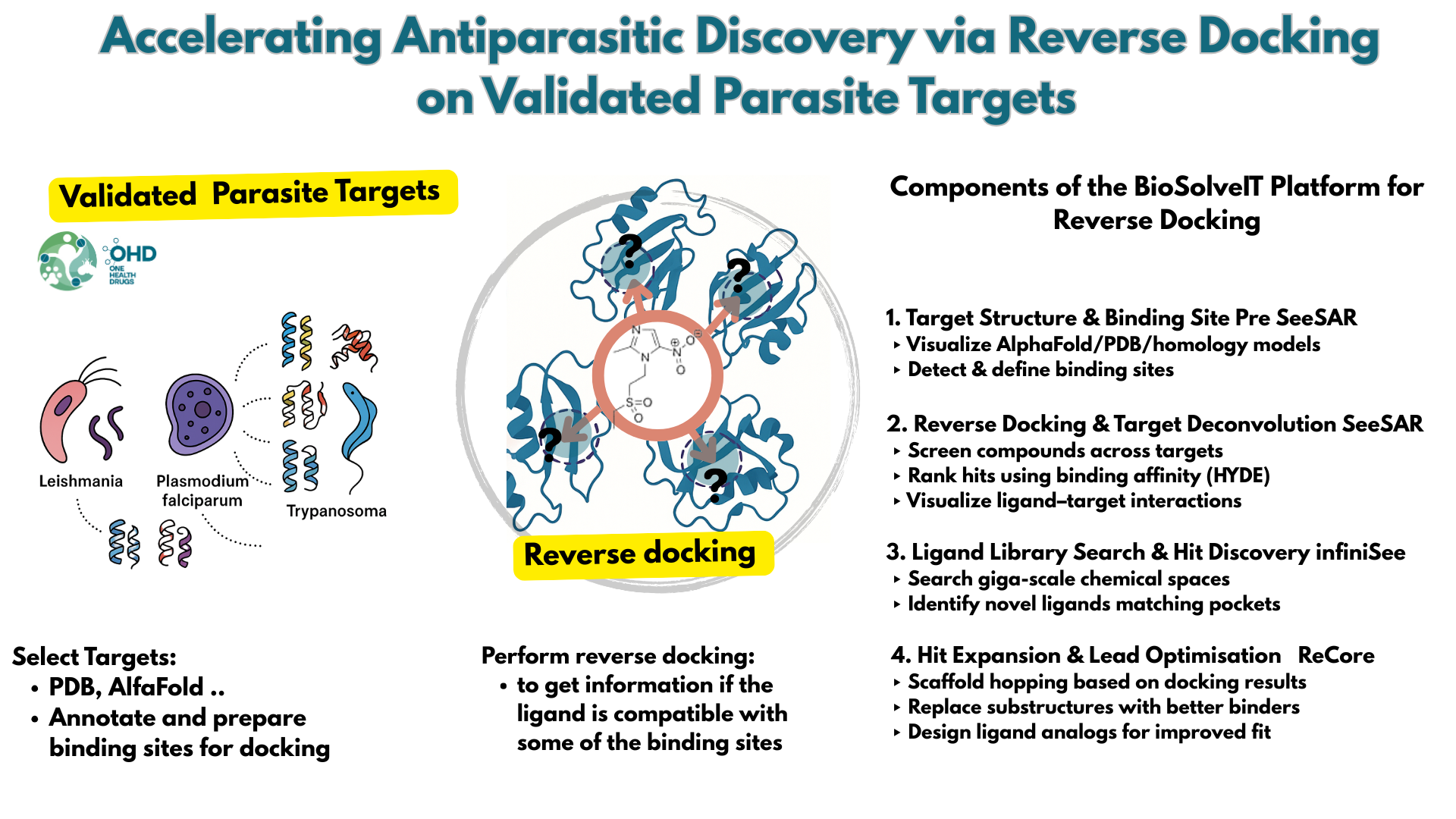
We propose to develop a structure-annotated panel of validated antiparasitic protein targets from Leishmania and Trypanosoma species for reverse docking applications. High-quality 3D models will be generated from AlphaFold, PDB, and homology modelling, with binding sites defined using structural alignments and ligand data.
BioSolveIT’s SeeSAR, infiniSee, and ReCore tools will explore target–ligand interactions across parasite proteomes. The COST CA21111 Action will guide target selection and ligand libraries, using compounds collected within its framework. The most promising ligands identified through reverse docking will be prioritised for experimental validation. This integrative approach supports target deconvolution, hit prioritisation, and antiparasitic drug discovery.
Črtomir intends to achieve the following milestones:
- Generate and annotate high-quality 3D structures of selected parasite targets.
- Perform reverse docking using BioSolveIT tools with COST CA21111 ligand libraries.
- Prioritise ligand–target pairs for experimental validation within the COST network.