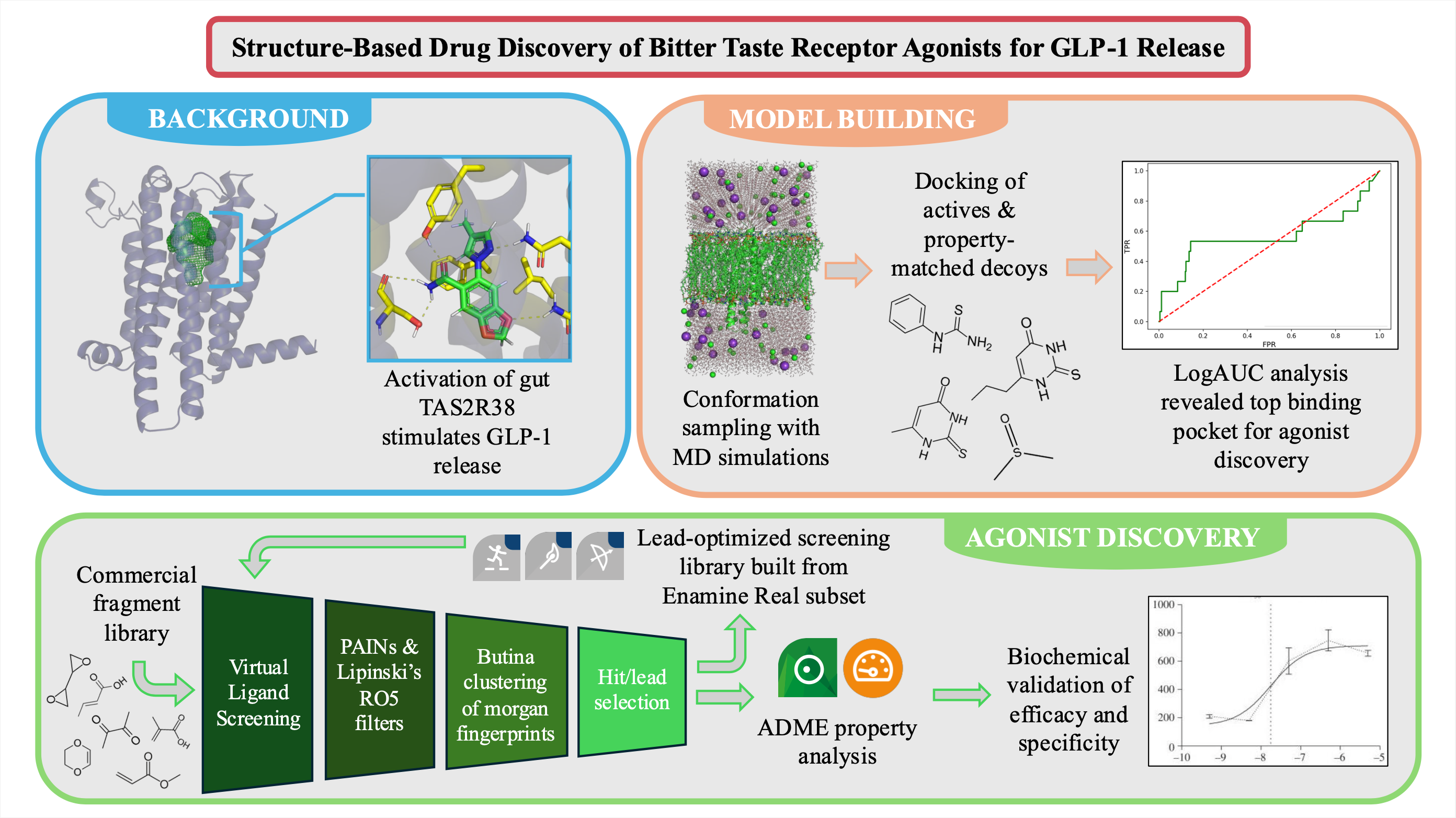
A TAS2R38 binding pocket model was successfully validated for enrichment of known agonists, enabling large-library fragment screening and analog library construction. Receptor conformations were sampled using Molecular Dynamics (MD) simulations, and ROC-logAUC analysis identified frames library screening. While initial AUC scores were modest, applying logAUC to emphasize early enrichment uncovered a strong candidate. This validated binding pocket, stemming from a holo MD simulation, was then used to screen a commercial library of fragment compounds. Lead compounds identified from the initial screen served as query compounds for the suite of BioSolveIT chemical space navigation programs to search the Enamine Real Space. This lead-optimized library is currently being screened against the receptor model. SeeSAR will be used to assess the pharmacokinetic properties of hits before in vitro efficacy validation.
After 3 months, Dylan has achieved the following milestones:
- A subset of the homology-based model of TAS2R38’s conformational space was sampled via AMBER MD simulation: 20 µs apo and 4 µs holo (agonist-bound). To maximize diversity, MD frames were clustered. 15 ligands with experimentally validated efficacy were docked against a background of property-matched decoys to each cluster representative. A set of AlphaFold2 models was compared, but a highly homologous modeling template followed by agonist-bound MD yielded the most promising binding pocket for screening large chemical spaces for agonists.
- A commercial, in-stock fragment library of 954,000 unique ligands was prepped to generate a screening space of roughly 1.2 million conformers. This library was docked against the validated binding pocket of TAS2R38 to identify promising agonist scaffolds. Ligands with a dock score of less than -8.5 kcal/mol were selected and filtered to remove PAINs substructures and Lipinski’s rule of 5 violators using rdkit. Remaining ligands were clustered using the Butina algorithm and Morgan fingerprints. Post processing, a diverse set of 111 lead TAS2R38 agonists was identified for further optimization.
- Lead compounds identified from the initial screening campaign, along with the 15 experimentally validated agonists, served as query molecules for FTrees, SpaceLight, and SpaceMACS chemical space exploration. Each seed molecule was used to search the 76.9-billion ligand Enamine Real space for purchasable analogs. These compounds were combined into a lead-optimized screening library of 94,500 ligands, which is currently being ranked based on dock scores against the validated TAS2R38 binding site. Post filtering and clustering of the top-ranking compounds, hits will be identified via manual binding pose inspection. Hits will be uploaded to the SeeSAR Analyzer to investigate ADME properties. Remaining hits will be purchased and have their efficacy and specificity validated in vitro via calcium imaging assays.