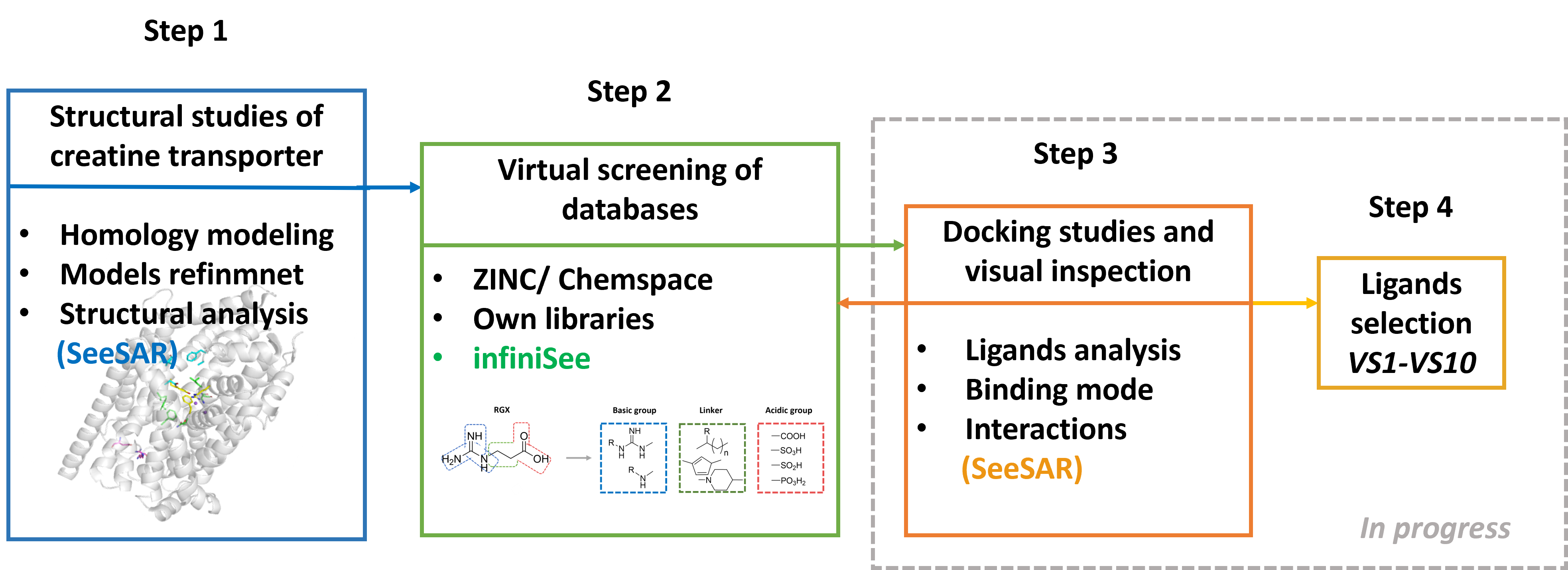
I established the structural basis for virtual screening of CT-1. Nearly 10,000 homology models were developed and validated in different conformational states, addressing the obstacle of the missing experimental structure. They were evaluated with scoring functions such as QMEAN, DopeScore, and Verify3D. The best models in outward-open, outward-occluded, inward-occluded, and inward-open states were selected for loop refinement. The next step involved analysis of differences in domains and residues between different conformations of CT-1. Visual inspection was performed in PyMOL and SeeSAR. Ligands are expected to bind to residues from the extracellular gate and the S1 site, located between TM1, TM3, TM6, and TM8. A major challenge was the limited number of known CT-1 ligands, which makes validation more difficult and increases the need for extended virtual screening in large chemical databases.
After 3 months, Dorota has achieved the following milestones:
- I analysed compounds with known activity against CT1 to access structural features important for biological activity. Ligands were grouped according to their chemical structure: presence/absence of acidic/basic groups, charge, aromatic rings, size of the linker and other properties connected with Rule of Five such as molecular weight, logP, number of hydrogen bond donors, hydrogen bond acceptors, to check which physicochemical properties are beneficial for transporter inhibition. Next, virtual screening (VS) of databases was conducted. VS was based on a substructure search of creatine and known CT-1 ligands. From the ZINC and Chemspace databases, approximately 10,000 compounds containing guanidine or amine groups linked to carboxyl, phosphate, sulfate were selected. Compounds with new scaffold appeard. Additionally, the engine infiniSee was implemented to identify near 6 000 compounds similar to creatine.
- Docking of compounds from the first screening has been initiated in Maestro, using multiple conformational states of CT-1 models. Docking grids were centered on key residues of the extracellular gate and the S1 site, which are expected to be critical for substrate recognition. Several compounds showed favorable orientations of guanidine and acidic moieties, forming contacts consistent with the proposed binding mode of creatine. Promising binding modes have therefore been observed, but the refinement to a final list of ~10 top candidates is still ongoing. This milestone will be completed using SeeSAR for additional docking, rescoring, and detailed visual interaction analysis, which will ensure reliable prioritization of compounds with reproducible and stable binding poses.
- Preliminary inspection of docking poses revealed interactions consistent with the creatine binding pattern, such as stabilization of the guanidine moiety near Cys144 and contacts with Gly73, Tyr148, and sodium ions. A full evaluation of the top 10 compounds with beneficial interactions is planned for the next months. These candidates will be analyzed and validated in SeeSAR, and extended with related purchasable analogs from BioSolveIT’s compound collections. This milestone is therefore in progress and will be finalized after refinement of binding mode analysis.