We have successfully completed our 1st milestone and working through the 2nd; BioSolveIT resources not only enabled this but expanded our typical CADD workflow. We’re also excited to share that our abstract was accepted at Canada’s largest chemistry conference – CSC2025, where we’ll talk about our findings and BioSolveIT tools (https://www.xcdsystem.com/cic/program/3NP1X0F/index.cfm?pgid=3066&sid=29948&abid=115159).
Over the past three months, we used Infinisee xREAL to develop libraries of potential 14-3-3ζ inhibitors, generating over 2 million compounds via Scaffold Hopper and Analog Hunter. These were docked and ranked using SeeSar and HYDE, identifying candidates with significantly better predicted binding than known inhibitors. Overall, the project is advancing well; however, the lack of data on compounds with high binding affinity for 14-3-3ζ is a challenge; that further underlines the necessity of this work; we aim that our data in coming months will pave the way.
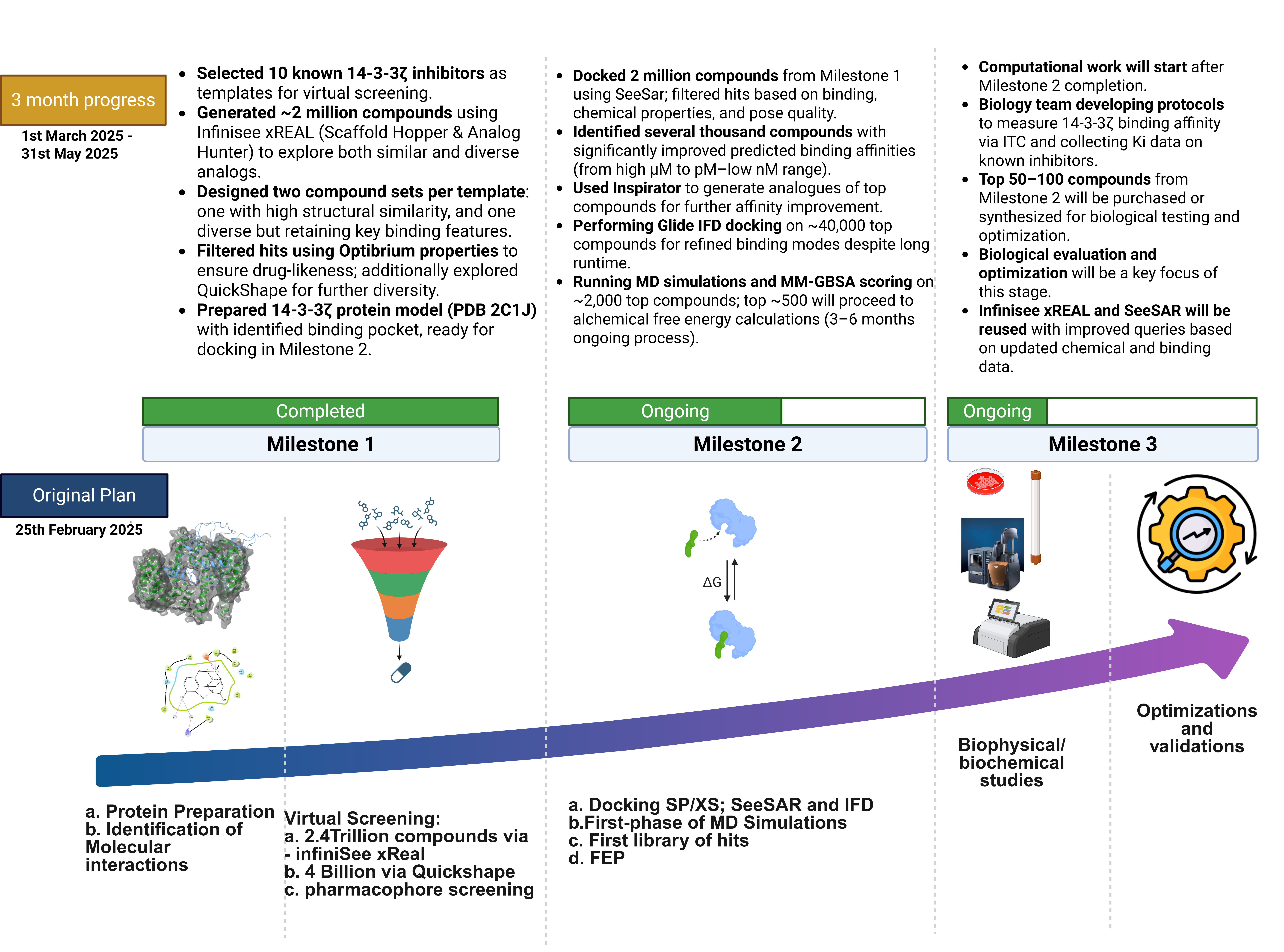
After 3 months, Samra has achieved the following milestones:
- Milestone 1 is successfully completed. We used 10 known inhibitors of 14-3-3ζ as query on Infinisee xREAL to build libraries of new potential inhibitors. Ihe vast chemical library in xREAL simplified the compound identification. We used scaffold hopper to identify similar compounds and analog hunter for diverse analogs, generating 200,000 compounds for each, totalling 2 million. We aimed to generate two sets per template: one with high structural similarity and one more diverse (lower similarity) but retaining key chemical features for binding. These were filtered using Optibrium properties to ensure drug likeness. We also used Quickshape on a prepared library, but it runs slower and used a 4 billion compound subset of REAL; hence, compound similarity was lower than with xREAL. Afterwards, we prepared the 14-3-3ζ protein model (PDB 2C1J, chosen for no missing parts), identified the binding pocket, and got it ready for milestone 2.
- We are halfway through milestone 2. First, we performed docking on the 2 million compounds from milestone 1 using SeeSar. Binding modes and affinities were compared with known inhibitors and filtered based on predicted binding, chemical properties (e.g., LogP), and poses (e.g., interactions, torsion quality), resulting in several thousand compounds with significantly improved predicted binding. Inspirator was also used on top compounds to derive analogues with better affinities. Overall, binding affinities improved from ~high µM for templates to pM–low nM for top hits. We are also performing docking with IFD in Glide for further refinement; due to long runtimes, only select top compounds are being docked (40K compounds). MD simulations are being run on top compounds from milestone (~2000 compounds), which will be scored using MM-GBSA. The top ~500 will advance to alchemical free energy calculations. This stage is ongoing and expected to take 3–6 months.
- Computationally, this milestone will begin once Milestone 2 is completed. Currently, our biology team is developing protocols to measure the binding affinity of 14-3-3ζ (via ITC) and is collecting Ki data for known inhibitors. This will ensure that once Milestone 2 yields our top hits, we can purchase the top 50–100 compounds from Enamine or synthesize them for biological evaluation and potential optimization. This will be a key part of the study. We will then reuse Infinisee xREAL and SeeSAR with updated queries based on improved chemical properties and binding affinities of 14-3-3ζ inhibitors.