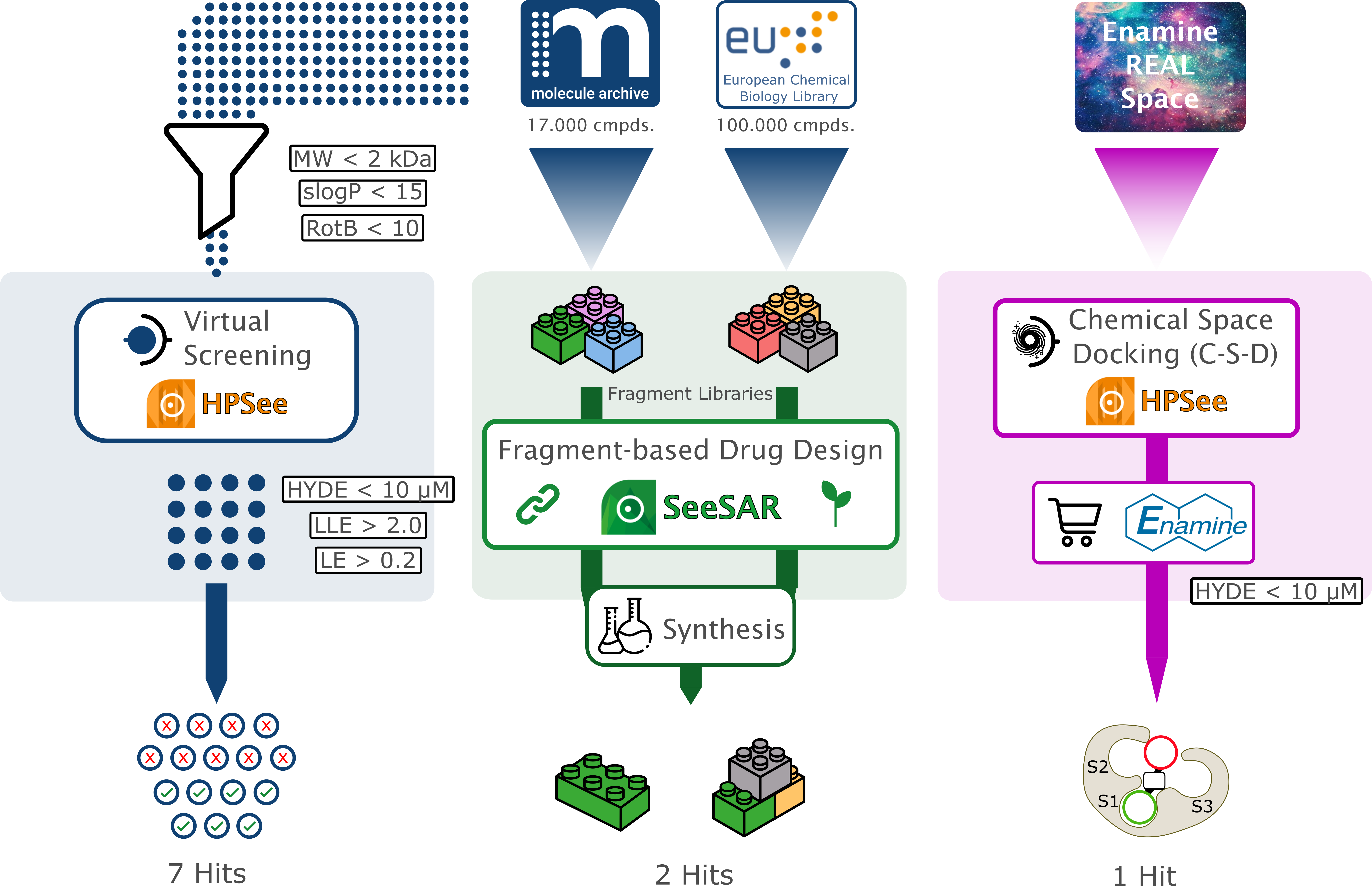
Our contribution to the Scientific Challenge Spring 2025 explored three strategies to discover novel inhibitors of the Zika virus protease using publicly available compounds from the Molecule Archive (MA) and EU-Openscreen (EUOS), and with the help of BiosolveIT software solutions SeeSAR and HPSee. Fragment libraries from MA and EUOS were initially subjected to crystallographic fragment screening (CFS) and yielded 23 hits (hit rate 14%) that served as the basis for subsequent design using SeeSAR. One fragment linkage was successfully synthesized. Crystal soaking of all obtained compounds from one design-make-test cycle yielded two new crystal structures with optimal exit vectors for further derivatization. A functional in-vitro protease assay demonstrated weak inhibition of Zika protease activity for one of the intermediates (20% at 50 µM).
The second strategy was a HPSee powered virtual screening of the MA, which currently contains approx. 17,000 substances from 44 international academic groups. Following an initial filtering step, the resulting 16,800 molecules were docked to the protease. HYDE scoring yielded 16 candidates for crystallographic validation. Of these, five were localized in the active site and two bound allosteric sites (hit rate 44%). In four cases, the experimental poses matched the highest-ranked docking pose. As the identified binders originated from research groups at KIT and the Universities of Greifswald and Mainz, a small synthesis consortium was formed to accelerate compound generation.
Chemical Space Docking (CSD) yielded two further compounds as high-scoring binders, one of which was obtained from Enamine and yielded another crystal structure after crystal soaking.
Combining fragment-based design via SeeSAR, and virtual screening as well as CSD using HPSee together with crystallographic validation, we obtained a total of 11 structurally confirmed Zika protease binders that now serve as a solid platform for structure-based optimization.
After 1 year, Christoph has achieved the following goals:
- Through fragment-based design approaches we obtained two crystallographically validated protease binders with one exhibiting weak inhibition in an in-vitro assay (20% at 50 µM). Using SeeSAR we designed several compounds, especially linking fragments of the S1 and S2 pocket. One promising compound (HYDE: 0.2–20 µM, LE: 0.3, LLE: 7.9) was successfully synthesized, however could not be localized within the binding site after crystal soaking. Instead, two synthetic intermediates revealed intriguing binding modes: Their central scaffold adopted a 180°-flipped orientation, still covering key interactions within the S1 pocket as predicted by SeeSAR. In this orientation, the moiety originally intended to address the S2 pocket forms an additional polar interaction with the S1 pocket. Consequently, the opposing part, initially designed to establish S1 interactions, is positioned towards the S2 pocket, providing an optimal exit vector for S2 pocket occupation and further hit optimization.
- Through virtual screening of the MA using HPSee, we identified seven crystal-soaking–validated binders of the Zika protease. From a total of 16,800 molecules, 16 candidates were selected for crystal soaking experiments based on HYDE scoring criteria (Affinity < 10 µM, LE > 0.2, LLE > 2.0). Seven compounds showed clear electron density in the crystal structures (44% hit rate), five of which were located within the active site. These active-site binders originated from three research groups at KIT, the University of Mainz, and the University of Greifswald, where fragment-based design and synthesis projects were subsequently initiated. This collaborative framework accelerates compound generation and systematic binding site exploration.
- We identified and crystallographically validated an additional Zika protease binder using Chemical Space Docking (Enamine REAL Space). Fragment-anchored chemical space docking yielded two highly ranked derivatives, one of which was readily available from Enamine and subjected to crystal soaking experiments. The crystal structure confirmed binding to the Zika protease and accurately reproduced the two key interactions within the S1 pocket as predicted by the docking algorithm. However, the molecular moiety intended to occupy the S2 pocket showed no electron density. Re-evaluation of the docking pose suggests that this is likely due to insufficient specific interactions with the S2 pocket. Nevertheless, the strong and well-defined S1 interactions render this CSD hit a promising starting point for further optimization and design.