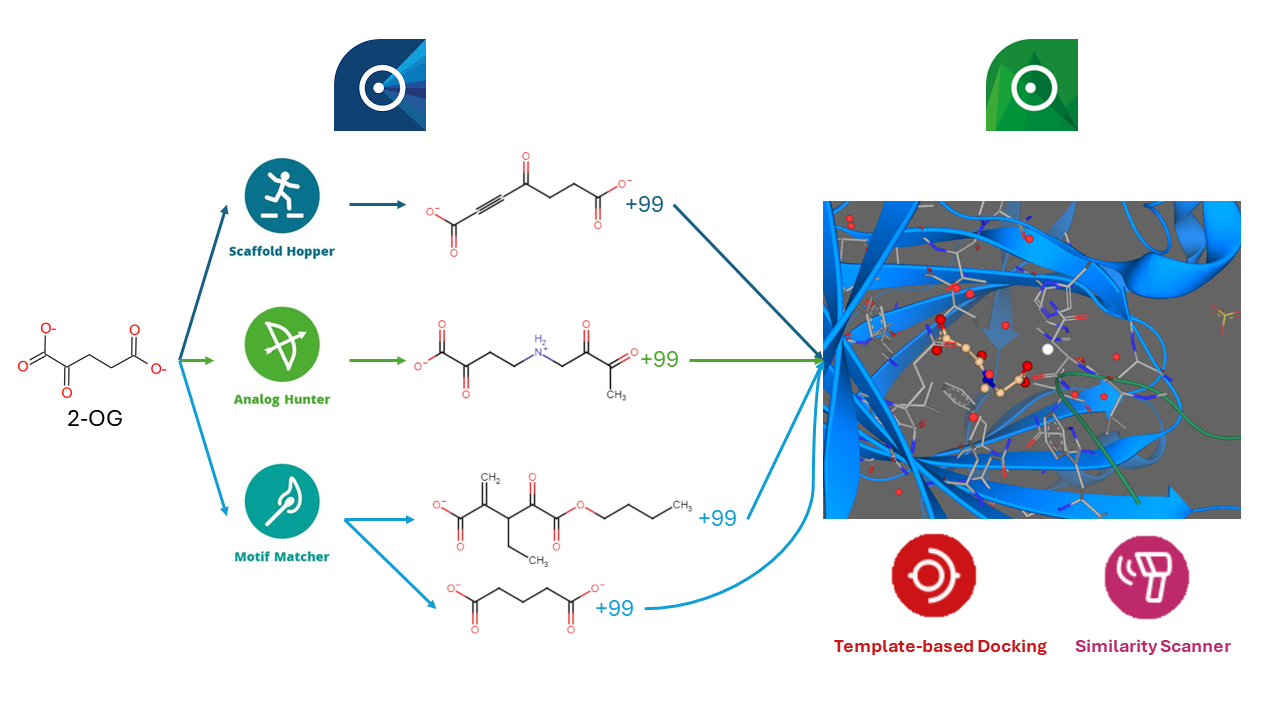
During the initial phase of this study, we utilized infiniSee software to identify novel molecules structurally akin to the natural ligand, 2-oxoglutarate. Packages employed included Scaffold Hopper, Analog Hunter, and Motif Matcher in both exact and MCS modes, collectively yielding 400 molecules for subsequent docking activities in Milestone 2. However, alignment and docking encountered challenges: the Similarity Scanner (FlexS) was unable to align several molecules—10 from FTrees, 80 from SpaceLight, 98 from SpaceMACS exact, and 56 from SpaceMACS MCS. Additionally, template-based docking (FlexX) lost some molecules during processing, hindering affinity predictions. These challenges necessitated further optimization of Milestones 1 and 2, emphasizing the need to refine processes to accommodate thousands of compounds. This optimization is critical for filtering and prioritizing molecules for future milestones, thus propelling the project toward identifying effective ligands.
After 3 months, Filipe has achieved the following milestones:
- We employed infiniSee software to pinpoint novel molecules resembling the natural ligand of the target protein, 2-oxoglutarate. Utilizing all available modules within the software, including Scaffold Hopper (FTrees), Analog Hunter (SpaceLight), and Motif Matcher (SpaceMACS) in both exact and MCS modes, we configured default settings to secure 100 results per method, aggregating 400 molecules for Milestone 2 docking. Future iterations will repeat this process on an expanded scale to generate thousands of potential candidates. Our initial analysis also uncovered several distinct scaffolds in the known ligand, providing valuable insights for potential future enhancements using this protocol.
- The alignment and docking phase presented numerous difficulties. The molecules derived from the similarity search were initially aligned against the original ligand using the Similarity Scanner (FlexS) under default settings. However, a significant number of these molecules could not be properly aligned, and subsequent attempts at template-based docking (FlexX) also proved unsuccessful. These issues led to failures in predicting the binding affinities of these new ligands. We are actively optimizing this workflow segment to overcome these obstacles effectively.
- This milestone has yet to commence as efforts are currently concentrated on refining the workflows in Milestones 1 and 2. Once these stages are optimized and applied to a broad set of compounds, we will proceed with designing new molecules using fragment-based de novo methods. Insights gathered from this milestone will be instrumental in filtering and refining the selection of molecules identified in the earlier milestones.