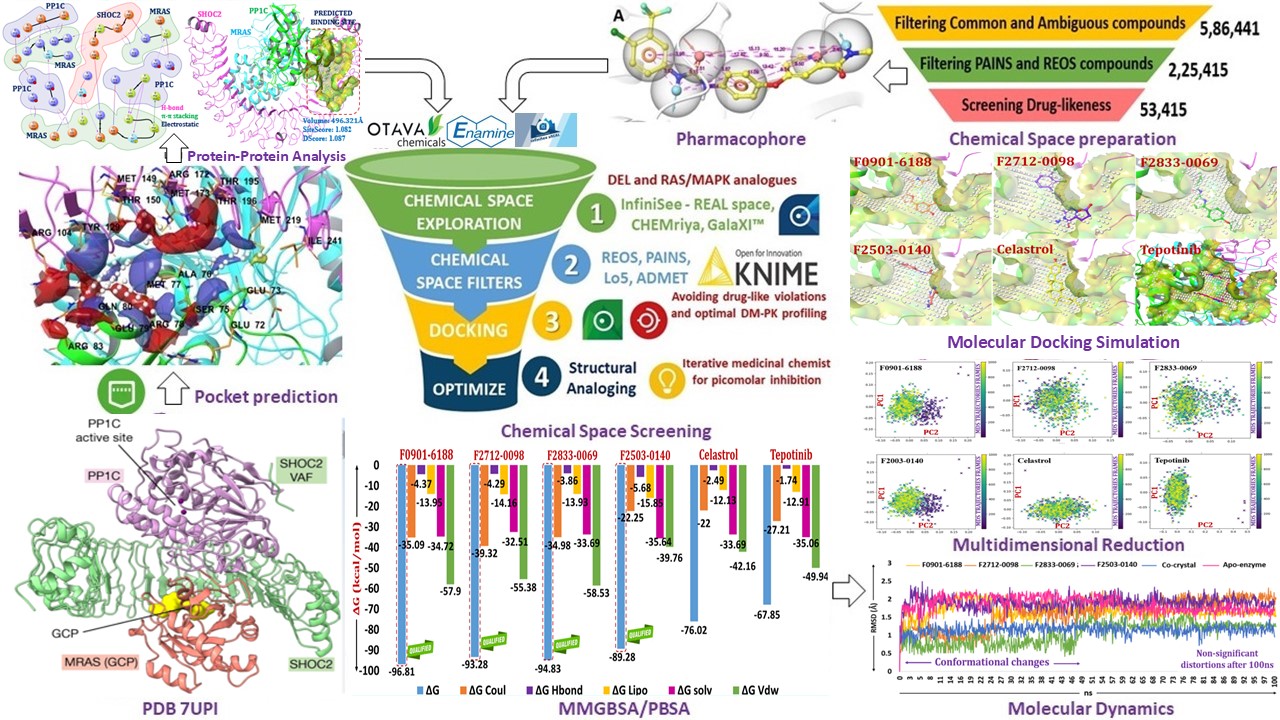
To begin with, I would like to express my sincere gratitude to BioSolveIT solutions for its remarkable speed, efficiency, and elegance, which made this year highly productive. My research focused on developing novel therapeutic strategies against Rasopathies by targeting both wild-type and mutant Ras proteins. Initially, I struggled to find a viable path to validate and advance my hypothesis. However, the Summer 2024 Scientific Challenge by BioSolveIT came my way to breathe life into my project. My hypothesis centered on targeting the SHOC2-MRAS-PP1C (SMP) ternary protein complex, originally studied by the Broad Institute of MIT and Harvard, and the Dana-Farber Cancer Institute. I aimed to identify putative leads capable of disrupting the SMP complex in both its wild-type and mutant forms, thereby alleviating Rasopathies. The SMP reliability was confirmed using AlphaFold, RoseTTAFold, and ProteinBERT. As the SMP complex exists in its apo form, druggable shallow and surface pockets were identified using DoGSiteScorer and validated through SiteMap. In-silico mutagenesis and known experimental mutations were mapped on the druggable pocket where the trimeric SMP interaction occurs. Using HYDE scoring and FlexX docking, we identified four key inhibitors, F0901-6188, F2712-0098, F2833-0069, and F2503-0140 fall between the picomolar to nanomolar range for both wild-type and mutant (M173I) complexes. Despite having their potency, these hits had solubility and metabolic hotspot challenges. These challenges were addressed through structural analoging and the incorporation of solubility-enhancing groups such as gem-dimethyl and monocyclic rings such as azetidine and oxetane. Derivatives of these hits demonstrated improvement in pharmacokinetic profiles without any violations. Further, frontier molecular orbital analysis and alchemical-steered simulations were used to gain deeper insights into molecular reactivity and confirm their binding potential. The optimized hits showed 200-fold higher affinity against mutants over the wild-type, with minimal conformational shifts, highlighting their promise as anti-Rasopathy chemotypes. Further biological evaluations are imperative, which are still underway to validate and translate them into successful therapeutic regimens.
After 1 year, Anuradha has achieved the following goals:
- Previously, we pioneered using the DEL library against the GTPase transducer MRAS protein. Here, we aimed to elucidate the binding mechanism of DEL with both wild-type and mutant forms of SMP complex. Key challenges included identifying selective binding residues, avoiding off-target engagement, and predicting composite active sites for chemical space screening. Using the Protein Editor module, we employed the MRAS protein's binding site as a template, producing an apo form of the SMP complex (PDB 7UPI) having low RMSD. Composite active sites were predicted using CASTp, SiteMap, and FastGrow, confirming a druggable pocket at the SHOC2-MRAS interface. Initially, the standard drugs Celastrol (SHOC2 inhibitor) and Tepotinib (MRAS inhibitor) were docked and validated using OpenBPMD yielding reliable pose. Based on the pharmacophore modeling, the curated chemical space was screened, and F0901-6188, F2712-0098, F2833-0069, and F2503-0140 had superior picomolar inhibition than standard drugs.
- The obtained hits F0901-6188, F2712-0098, F2833-0069, and F2503-0140 were assessed for ADMET analysis using Optibrium StarDrop, QikProp, ADMET-AI, and pk/tox-CSM (University of Queensland). The analysis revealed have lack of interactions with M173I projecting towards the growing space and modest Optibrium properties. The structural analog of these four hits was performed using FragGrow, SpaceLight, SpaceMACS, and Colibri. The newly designed series was predicted for false positive readout using PAINS (Pan Assay Interference), and REOS (Rapid Elimination of Swill) using DataWarrior and KNIME workflow, and further trimmed. Amongst the optimized series, F2503-0140A and F2712-0098C had a picomolar to nanomolar inhibition boundary and demonstrated 200-fold higher affinity than the standard drugs (Celastrol and Tepotinib) without any ADMET violations. Further, network analysis has been established with various Ras/Raf isomers as a multi-modal strategy for MAPK signalling inhibition.
- The cause of the mutant conferring rasopathies syndrome is due to the M173I. The wild-type residue Met173 has been mutated to Ile173 situated at the SHOC2 monomer, and the structure has been prepared using the protein editor module. The F2503-0140A and F2712-0098C have been probed at the orthosteric envelope to assess the binding affinity of M173I using the per-residue energy decomposition method. Further, the docked mutant complexes were driven for unbiased conventional molecular dynamics and steered molecular dynamics using AmberMD. The overall MMGBSA/PBSA of F2503-0140A and F2712-0098C exhibited higher binding affinity with the mutant Ile173 over the wild-type Met173 residue. The conformational analysis of these best complexes was investigated using the multidimensional reduction using MODE-TASK. The extracted trajectory frames were analyzed for the PCA and demonstrated to have non-significant deviations of the mutant complexes.