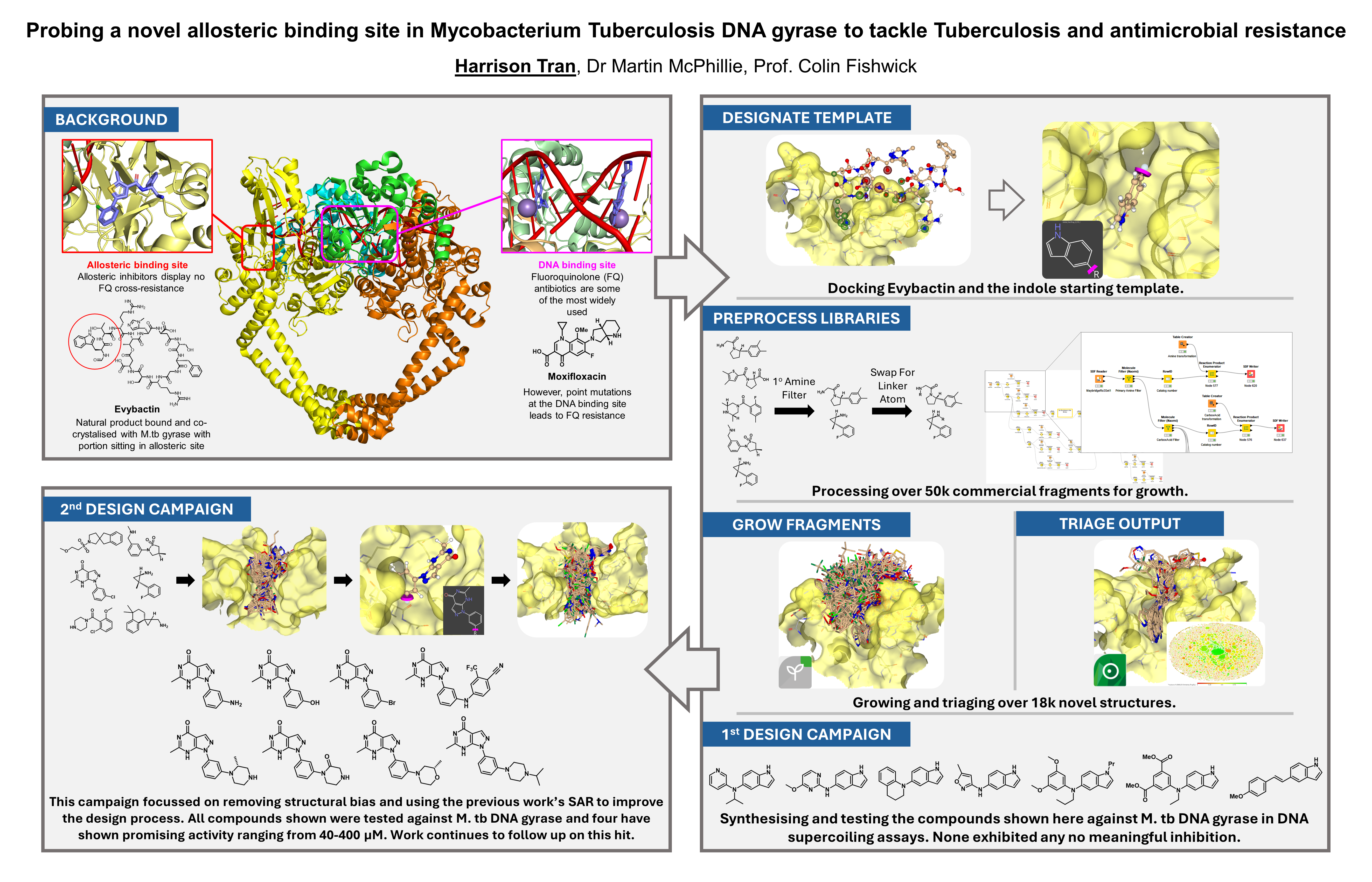
Over the last twelve months I’ve been using SeeSAR and its FastGrow module to try and achieve my goal of designing novel allosteric inhibitors of M. tb DNA gyrase. To date, BioSolveIT’s software has helped to produce over eighteen thousand new scaffolds, accomplished using FastGrow’s fragment growth in combination with more than 50k commercially available fragments. A total of 15 compounds have been synthesised from these designs of which four have potential activity. During the last year two major bodies of work have been conducted using BioSolveIT’s software suite, the first was inspired by the natural product Evybactin while the second completely de novo. In both instances we were able to produce a wide variety of structurally diverse and interesting molecules tailored to the target site.
One of the main goals of this work was to apply fragment-based drug discovery and computational fragment growth to a structure-based approach, in turn creating an iterative non-specific workflow. Thanks to SeeSAR’s docking abilities and ease of use this was accomplished by the second design campaign, with it using the lessons learnt and SAR previously explored to inform its design. It was this process that saw estimated binding affinities dropping from µM to sub-nM and the production of the first compounds to show biological activity. Moving from the natural product-inspired indole template to the commercially-inspired pyrazolopyrimidinone not only saw the start of potency but also an improvement in synthetic tractability. Having learnt a lot from the first design attempt, the second was much smoother with all the previous fragment processing I had previously undertaken applied to the new core with FastGrow very easily. At present synthesis still continues with further compounds being made to explore this new scaffold and expand SAR, using the µM hits discovered to guide the process.
In summary, during the BioSolveIT challenge the software provided to me has helped me take a massive leap forward in my PhD research. Allowing me to explore a brand-new approach with none of the risk and granting me access to cutting-edge software. My goal of designing novel allosteric inhibitors is now one step closer to being realised thanks to SeeSAR and FastGrow’s ease of use and wide applicability. Using BioSolveIT’s suite of accessible software led to nearly 20,000 novel designs, the synthesis of 15 novel compounds and 4 promising µM hits.
After 1 year, Harry has achieved the following goals:
- My first goal was to use SeeSAR and FastGrow to enable the de novo design of novel allosteric binders of M. tb DNA gyrase. I achieved this aim across two different design campaigns: the first inspired by Evybactin (a known allosteric inhibitor), the second using SeeSAR’s docking module to start from scratch, aiming to eliminate any previous structural biases. This was done by taking a selection of diverse commercial fragment libraries, processing them into growth substrates, and then combining them with template molecules docked within the target. The workflow was designed to be chemistry-driven hoping to ensure synthetically accessible outputs. For instance, KNIME was used to filter and create linking partners from specific structural motifs, like amines or hydroxy groups. These were then paired with the other appropriate functionality to simulate typical toolbox reactions. As a result over 18k novel scaffolds were designed, tailored and grown specifically for this allosteric site.
- My second goal was the chemical synthesis and biological evaluation of the novel molecules I’d designed. Over the 12 months I produced and tested over 15 novel compounds, of which four exhibited promising activity within the micromolar range. Efforts towards this aim started with the triaging and analysis of the diverse grown fragments. At this stage in both campaigns decisions were primarily made based on the estimated binding affinities generated by SeeSAR, synthetic accessibility and overall structural similarity, to avoid synthesising scaffolds with similar 3D-shapes. This, given the design of the workflow, produced shortlists of very interesting molecules. After selection came the synthesis, as the scaffolds produced were created to be diverse, so was their chemistry and in some instances required extensive optimisation. However, these issues were eventually solved and 18 novel compounds were produced and tested against M. tb DNA gyrase in a biochemical supercoiling assay.
- My final goal was to use biological data I’d collected to inform further design. Mentioned throughout has been the pursuit of two design campaigns, this aim was achieved upon the start of the second. The first attempt was inspired by Evybactin, using an indole from its tryptophan residue as the design template. Following synthesis and testing of some designed molecules it was clear that the scaffolds produced had little-to-no activity. This suggested that the indole motif wasn’t an effective starting point and another fragment might be better. Learning from the previous compounds, I started the process from scratch by docking commercial fragments, selecting the best, growing it and selecting outputs distinct from the first campaign. SeeSAR’s predicted binding affinity went from µM to sub-nM and was reflected in biological testing, with 4 of the new compounds showing activity. This outcome achieved my goal and highlighting FastGrow’s power in an iterative structure-based approach.