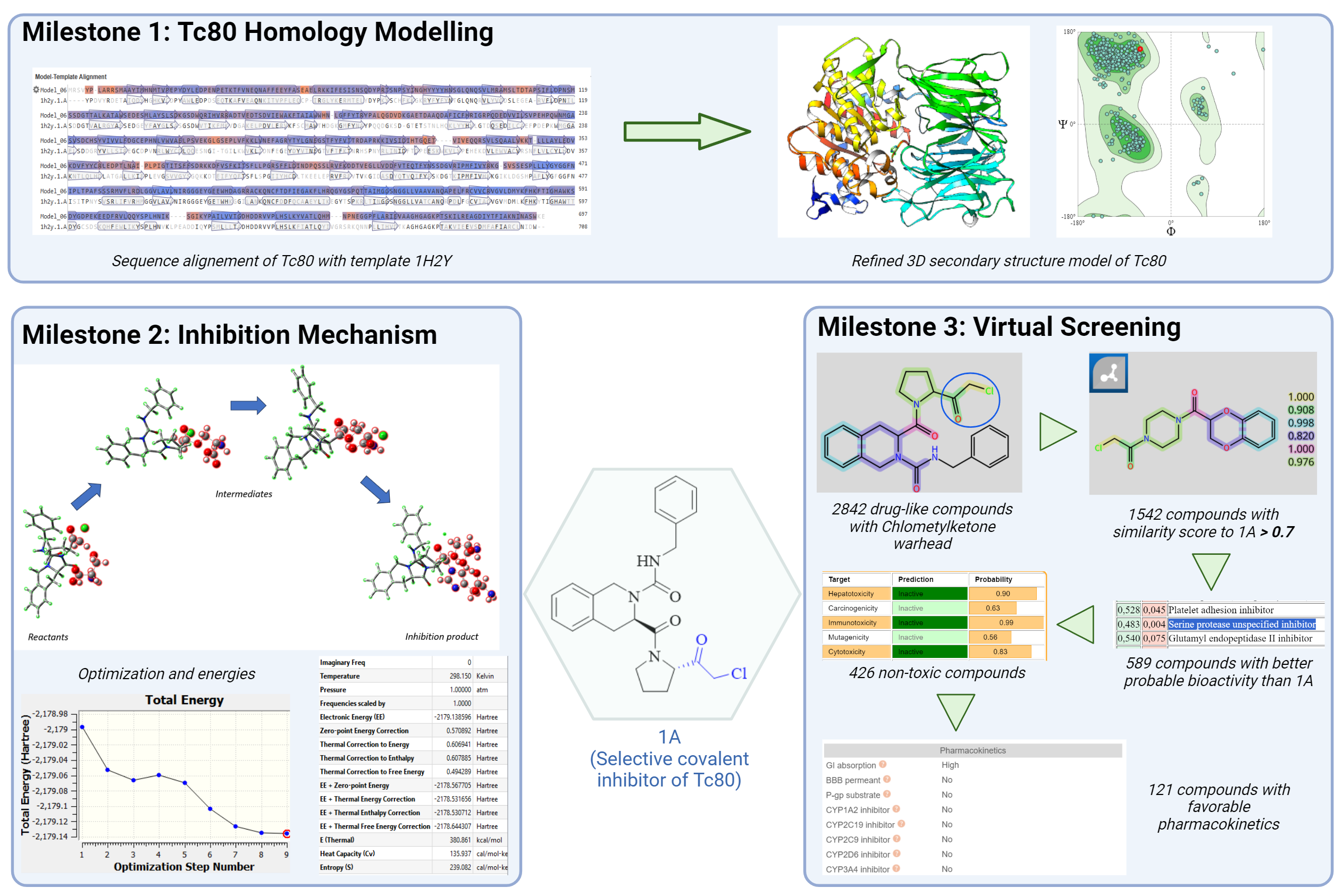
The first obstacle was the unavailability of the crystallized structure of Tc80. To overcome this, the 3D secondary structure of Tc80 was built by homology modeling methodology. We chose different templates for the construction of the 3D secondary structure model of Tc80 using the Swiss Model. The best model was refined using the GalaxyWeb server and validated using the SAVESv6.0. On the other hand, we wanted to find compounds with better pharmacokinetic properties than 1A through the use of computational tools, therefore, we built a library of 2842 drug-like compounds with chloromethyl warheads extracted from commercial libraries. Then, these compounds were filtered using FTrees, PASSONLINE, ProTox, and SwissADME resulting in 121 compounds. The last obstacle we had to overcome was the lack of studies on the mechanism of chloromethyl ketone inhibitors against Tc80. We employed the Density Functional Theory (DFT) and obtained stable intermediates.
After 3 months, Ivan has achieved the following milestones:
- The sequence of T.cruzi oligopeptidase Tc80 was obtained from the NCBI database. The templates to construct the 3D secondary structure model of Tc80 were chosen from the Swiss-Model web server. The 10 best templates were chosen to build the 3D model of Tc80. These models were validated with ERRAT, VERIFY 3D, PROCHECK, and Ramachandran plots. The best model was built with the prolyl oligopeptidase from a porcine brain X-ray structural template (PDB ID:1H2Y) and 44.1% sequence identity. This constructed model was refined using the GalaxyWeb server. After the refinement, the validation parameters were enhanced, indicating a more reliable 3D structure of Tc80 (more than 96% of residues in the favored Ramachandran plot). This 3D model of Tc80 will be used to perform the covalent docking with SeeSAR software.
- The inhibition mechanism was carried out in basis set B3LYP/3-21g and B3LYP/6-311g (d,p) using Gaussian software because they were the most accurate and suitable for geometry optimization, where we wanted to find the stable molecular structure, bond lengths, and angles adequate. On the other hand, B3LYP/6-311G(d,p) is appropriate for medium-sized molecules where an optimum balance between accuracy and computational cost is desired. We obtained the structure of stable transition states, reactants, and products. The mechanism begins with the serine activated undergoing an SN2 reaction to form a hemiacetal which was corroborated using Intrinsic Reaction Coordinate. Then the oxygen of tetrahedral hemiacetal negatively charged showed a preference for the carbon bonded with chlorine, thus giving rise to the formation of the epoxid ring. Finally, the histidine enables the bond to break out of the epoxid ring and link up to the inhibitor.
- 76,496 covalent compounds were extracted from Life Chemicals, ChemDiv, Enamine, ChemSpace, and CovalentInDB. The warhead (chlomethylketone) screening was performed with DataWarrior software obtaining a library of 2842 unique molecules. From these compounds, 1542 resulted in a similarity score relating to 1A above 0.7 using FTrees software. The probability of the Serine protease unspecified inhibitor activity of these compounds was calculated with the PASSONLINE web server using 1A probability activity value as a threshold, obtaining 589 compounds with better probability activity. Toxicity filtering was applied using the ProTox web server to avoid potential hepatotoxicity, immunotoxicity, mutagenicity, and cytotoxicity, obtaining 426 compounds. Pharmacokinetic filtering was applied using the SwissADME web server searching for compounds with high gastrointestinal absorption, avoiding PAINS alerts and activity with the cytochrome P450 family resulting in 121 remaining compounds.