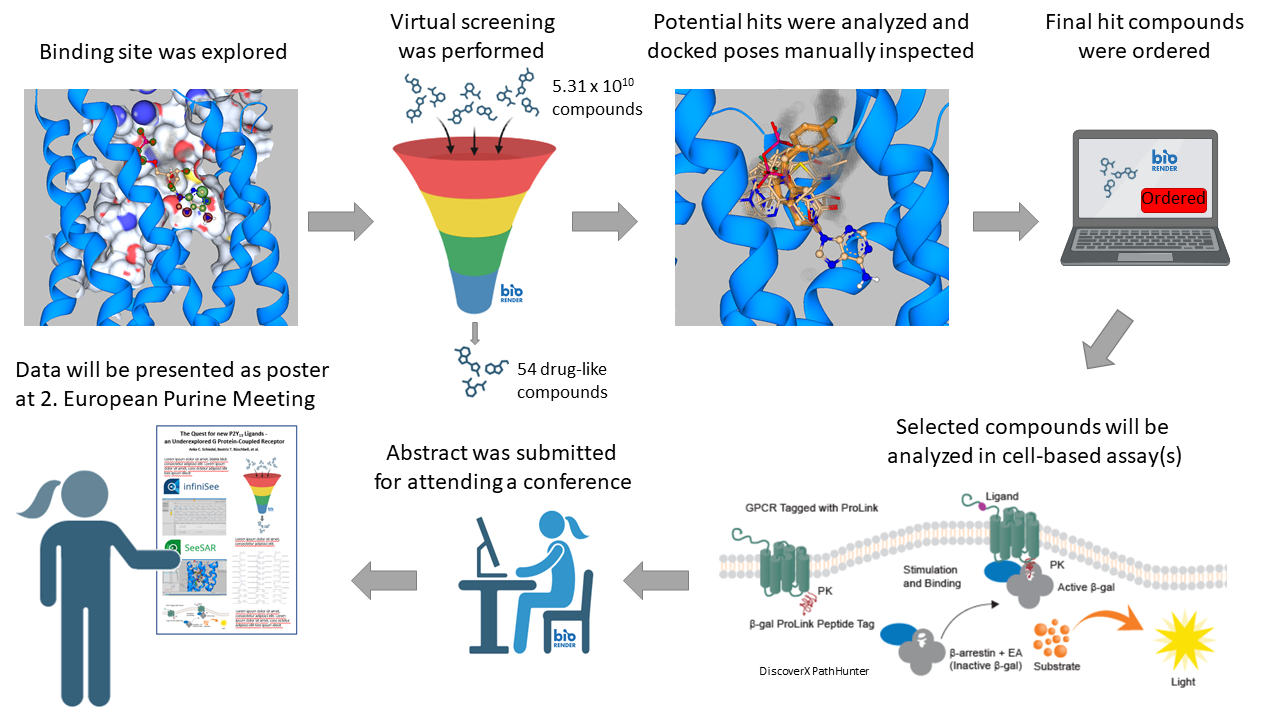
The overall goal of the P2Y13 project to find new ligands was reached. We were able to identify 52 drug-like compounds with 32 different scaffolds which could be successfully docked into P2Y12 structures and also into P2Y12 mutants mimicking the binding site of P2Y13. Docking into P2Y13 models was also performed, but the initial problem with too many clashes of the docked molecules could not be overcome so far. The P2Y13 Alpha-fold model did not result in better docking poses either. Therefore, we built two models of the P2Y12 receptor based on the structure 4PXZ1 which were modified in the binding pocket to mimic the binding site of P2Y13. P2Y12 is the closest relative of P2Y13 with a crystal structure. P2Y12 and P2Y13 bind with high affinity 2MeSADP, the co-crystallized ligand of the structure 4PXZ. The differences for the binding of 2MeSADP are two amino acids, F106P2Y12/E125P2Y13 and T163P2Y12/S182P2Y13 and for AZD1283 a third amino acid, L155P2Y12/I174P2Y13 was different.2 InfiniSee was used for the virtual screening approaches of several compounds. First, we used the Scaffold Hopper tool and second, we searched with the Analog Hunter tool. We searched 5.31 x 1010 compounds from the REAL and Freedom Spaces. The hits of each query were analyzed towards drug- and/or lead-likeness. The virtual screening approaches yielded 52 drug-like compounds with 32 different scaffolds. 13 compounds were found in the Freedom Space and 39 in the REAL Space. 45 of the 52 final hits were also lead-like. The individual subsets were then docked into both mutant receptors and best poses were taken and carefully inspected. A final set of seven compounds was ordered from Enamine. As backup we plan to order the best hits obtained with searching the Freedom Space.
In addition to the virtual screening and docking project within the challenge project we optimized the -arrestin assay in the lab, which will be used for the pharmacological characterization of the ordered compounds. This assay was already validated and published.3 A second assay, the TRUPATH assay is tested for suitability for P2Y13 at the moment. Both assays are already set up for the P2Y12 receptor, which will be used as control for the potential P2Y13 ligand. Since the binding sites are very similar it is expected that the compounds should also show an effect at the P2Y12 receptor.
The results of the virtual screening and docking approach using InfiniSee and SeeSAR have been submitted as abstract for the upcoming 2. European Purine meeting held in Ferrara (Italy) from September 4-6th. If accepted it will be presented as poster.
References: 1 Zhang, et al. (2014) Nature 509: 119-122;2 Pérez-Sen et al. (2017) Adv Exp Med Biol - Prot Rev 19: 139–168; 3 Zhang et al. (2024) Nat. Chem. 16: 249–258
After 1 year, Anke has achieved the following goals:
- Virtual screening was successful and led to drug-like compounds which could be docked. Based on the results of milestone 1 from the 3-month report we inspected the docked poses of the virtual screening runs with cangrelor and 2MeSADP. Due to persistent clashes we decided to repeat the docking using the above described P2Y12 binding-site mutants. Further compounds were also used for virtual screening with the Scaffold Hopper and Analog Hunter. Especially the hits based on MRS2211 led to interesting compounds. Compounds were analyzed with the Analyzer tool in InfiniSee and only compounds which were drug-like and/or lead-like were considered for docking. All final sets were docked using SeeSAR and best poses were taken. Standard docking as well as template-based docking was performed and hits were merged in the end. After manual inspection and discussion with a chemist compounds were selected for ordering.
- The fragment-based approach was applied to the known ligands 2MeSADP, cangrelor and MRS2211. Structures were analysed in the Inspirator mode of SeeSAR and modified to either replace the phosphate groups and/or sugar moiety or the nitro group in MRS2211. Some of the proposed compounds seem to have improved potency. After pharmacological testing of the hits from the virtual screening the fragment-based approach will be repeated with the final hits and proposed compounds will be discussed with our in-house chemists.
- FastGrow was used so far only with standard compounds and tried with some of the final hits from virtual screening. After testing of the hit compounds FastGrow will be applied to the new ligands. Fast Grow will also be applied to compounds we hope to find in a second screening run of a new collection of our in-house proprietary compound library which will be started mid June.