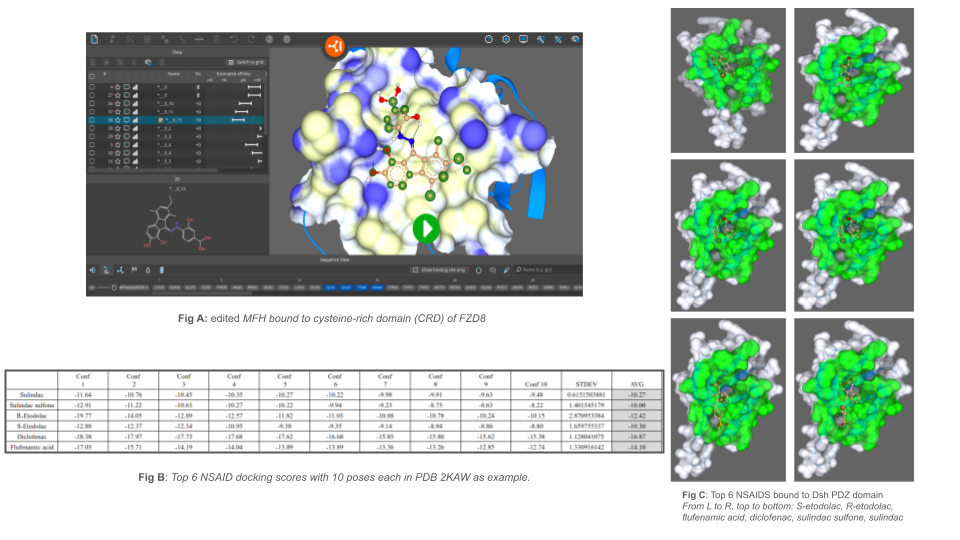
Over the past 12 months, we thoroughly evaluated the potential of small molecule “MFH” which is previously known to bind to the cysteine-rich domain (CRD) of Frizzled8. Using SeeSAR, we analyzed the properties of the amino acids in the pocket as well as the ligand properties, and tried to make edits to the molecule accordingly in order to increase the affinity and make more strong interactions. Unfortunately, the pocket itself is (a) very shallow and (b) made up largely of hydrophobic amino acids, and so the molecule interacts with the pocket using largely hydrophobic interactions. As a result, we were unable to make dramatic changes to improve the affinity, as only bulky hydrophobic groups would produce slight increases in affinity. Upon further exploration of the MFH molecule, we did discover that MFH potentially has specificity to Frizzled8 over other Frizzled isotypes due to other Frizzleds having a structural hump inside the same pocket that would prevent MFH from binding.
As a result, we decided to take a slight detour and work on Disheveled (Dsh), a protein that is just downstream in the same biochemical pathway when Wnt binds to Frizzled and LRP5/6. Based on previous research that sulindac, an NSAID, binds to the PDZ domain of Dsh, we used SeeSAR as well as FlexX to screen 35 NSAIDs in the PDZ pocket. From those, we identified the top contenders: sulindac, sulindac sulfone, S and R etodolac, diclofenac, and flufenamic acid. We are currently actively working on screening them in vitro in 293T cells as well as editing the top molecules in silico to see if we can produce an improved small molecule.
After 1 year, Margaret has achieved the following goals:
- The first important goal we reached during the last 12 months was that we maximized the binding potential of MFH to the CRD of Fzd8. We achieved this goal by using SeeSAR’s 3D modeling and surface rendering capabilities to analyze the protein’s CRD pocket and the way the molecule fits into the pocket. We noticed that the pocket is shallow and hydrophobic, and that the molecule mainly interacts with it using Van der Waals (VDW) interactions. In order to take advantage of the properties available, we used the Molecule Editor function in SeeSAR to make changes to MFH that we thought would increase the affinity, such as methyl groups and hydrophobic rings. However, due to the shallow and small nature of the pocket, the molecule MFH fit very snugly into the pocket, so any bulky edits were likely to shift the molecule to an undesirable position further away from the pocket. Regardless, we still improved the affinity as best as possible (Fig A).
- The second important goal we reached during the last 12 months was we screened a variety of NSAIDs in the PDZ domain of Disheveled. When we shifted our attention towards the downstream protein Dsh, we noticed that previous research demonstrated sulindac binds to PDZ of Dsh. On top of that, there are current clinical trials ongoing for ketorolac on the Wnt pathway in Retinopathy of Prematurity, a condition that is becoming more common as more premature babies survive with improved technology. Because ketorolac is an NSAID that shares similar size and groups as other NSAIDs in the same class of drugs, our project expanded interest into efficacies of other NSAIDs. We used FlexX and SeeSAR to screen 35 different NSAIDs for their binding ability to 20 different PDB codes of the PDZ domain of Dsh (Fig B - 2KAW as example). These included screenings for not only other acetic acid derivatives besides ketorolac, but also salicylates, as well as propionic acid and anthranilic acid derivatives.
- The third important goal we reached during the last 12 months was that we identified the top contenders and are working towards in vitro testing and in silico editing. We have identified 1) diclofenac, 2) R-etodolac, and 3) flufenamic acid as the top contenders based on docking score/affinity as well as binding position visually. We enhanced the accuracy of binding such that specific residues from sulindac’s original pose to Dsh were retained across all of the small molecules. For this, we used a pharmacophore to fix groups like the carboxymethyl. The compounds have been ordered and we are currently waiting for them to come in so we can conduct in vitro assays on 293T cells. While we are waiting, we are currently trying to edit each of the top scoring NSAIDs to possibly make a better binder (Fig C).