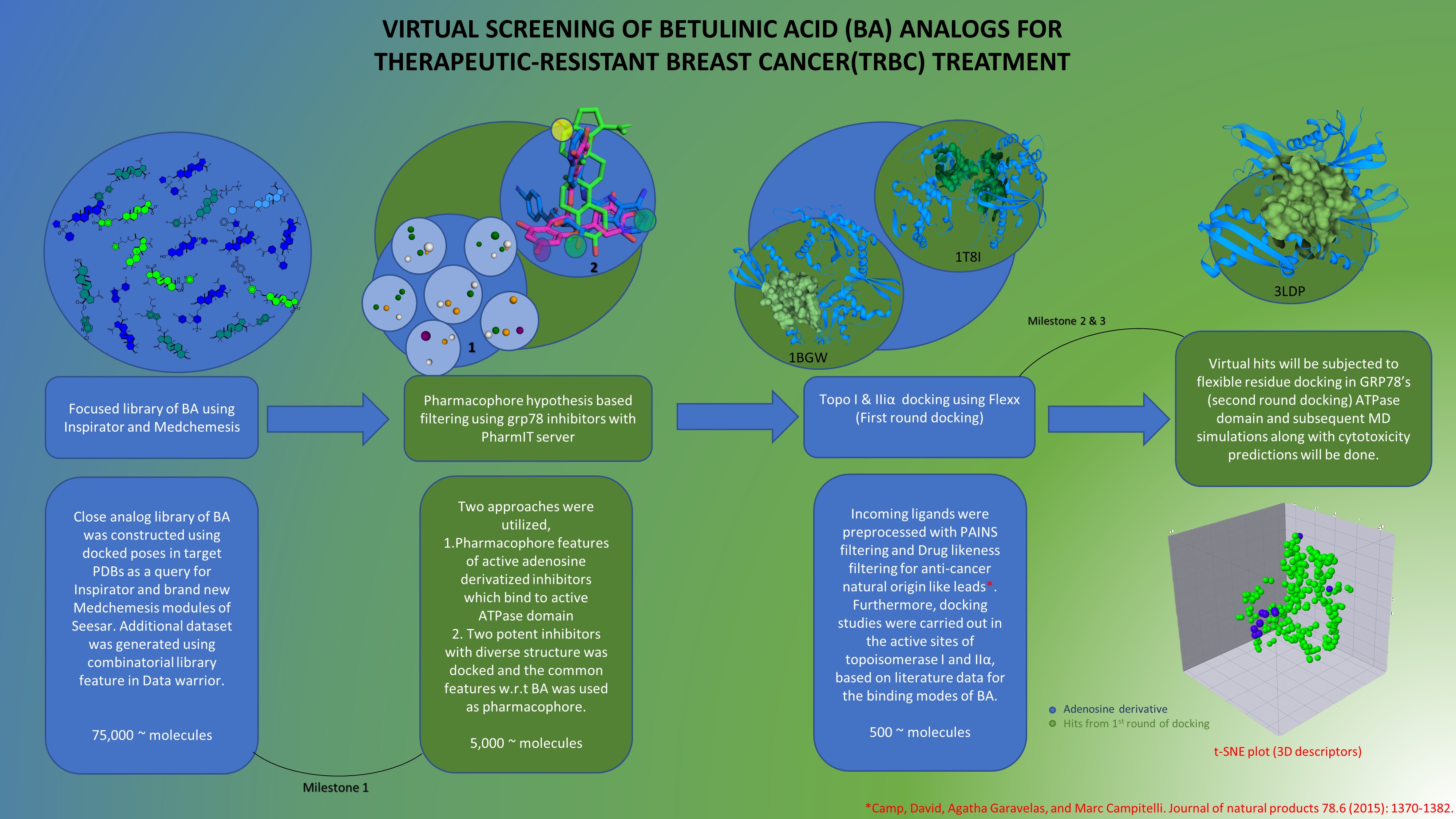
The close focused analog search using Seesar & Swiss similarity, along with all the supplied catalog libraries, did not yield satisfactory results. A combinatorial library for betulinic acid derivatives was created using literature-derived chemical transformations and building blocks from Chemspace, Enamine, and Zinc databases. The Inspirator module modified the side chain of BA in 3 target PDBs, resulting in a dataset capturing geometric features of the active sites. Furthermore, BA derivative space was expanded using new Medchemesis feature. Library of ~75k was built by subsequent filtering & processing to remove unwanted atoms. Pharmacophore models were generated using two approaches 1. pharmacophoric feature of potent adenosine derivatives with subsequent validation with decoys 2. Docked poses of Adenine derivative inhibitor & EGCG was used to derive common features to explore subdomain IIa, IIb & Ia. Docking was carried out in on topoI and IIa using knime interface.
After 3 months, Sagar has achieved the following milestones:
- Focused library was designed using Seesar's Inspirator and brand new medchemesis feature to generate dataset from three target structures and using all combinations of supplied fragment libraries in seesar module. This library was further expanded using literature derived chemical transformation of BA yielding potent anti-cancer molecules. Two approaches were utilized to filter the compounds based on grp78 inhibitors 1. Adenine derived grp78 inhibitors were searched using chembl and the dataset was imported and aligned on the most potent adenine inhibitor 3GO active conformer and common features were extracted. 2. To include diversity and explore pockets not explored by the adenine-based ligands EGCG was used as it is having potent grp78 inhibition(0.7µM) also found to be a topoisomerase poison thus supporting the hypothesis for dual targeting. The docked conformations of the three ligands were then used to generate pharmacophore model to filter the focused library dataset.
- Virtual hits from two multiple pharmacophore hypothesis were subjected to rule-based filtering for drug likeness of natural origin anticancer lead compounds retrieved from literature. Further PAINS filtering was done to remove these moieties generating false hits. First round of docking was applied on preprocessed topo I and IIa PDBs, the binding modes of BA in these proteins was derived from literature and subsequently optimal binding poses were generated by docking BA, later on this was used as the active site for virtual screening in both the targets. Docking was conducted using FlexX in knime interface on a personal laptop (I3, 8GB ram) with 5 conformers to reduce the time of computation. BA acids are topo catalytic inhibitors, such that BA analogs interacting strongly with the catalytic domains on both the sides were selected. Further docked hits from both targets were filtered with max threshold of -15 Kcal/mol. Further second round of docking and filtering will be conducted.
- Yet to be achieved.