The human autophagy system is an important pathway for the degradation of misfolded proteins and organelles. However, approaches to target the membrane-bound autophagy modifiers (LC3/GABARAP proteins, also known as Atg8 proteins) responsible for cargo recruitment have been largely unsuccessful for decades. Therefore, our goal was to identify non-covalent small molecule ligands targeting the main binding site of LC3A (called LIR docking site (LDS)) as potential tools for autophagy modulation and targeted protein degradation (TPD).
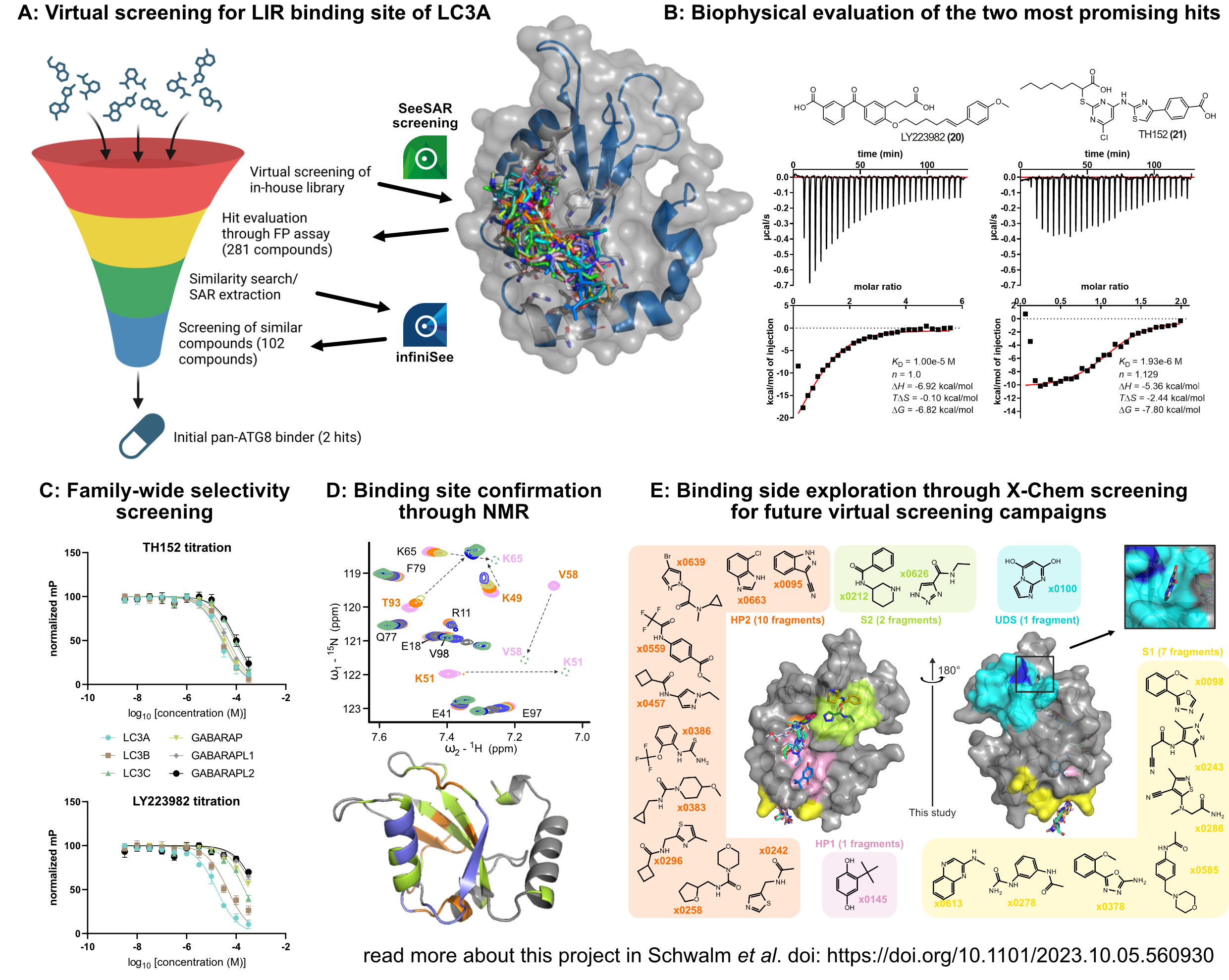
To identify new ligands, we used SeeSAR to virtually screen our in-house library, followed by biophysical pre-screening (using fluorescence polarisation) and a second iteration of screening using infiniSee similarity search on all validated binders (A). By using this pipeline, we were able to identify two non-covalent small molecules with affinities of 10 and 2 µM (currently the most potent small molecules targeting LC3/GABARAP) (B). Furthermore, we tested the compounds for selectivity within the whole protein family (C) and confirmed the binding site to the virtually screened LDS using in-solution NMR spectroscopy (D). Finally, we used X-ray crystallography to experimentally identify additional binding sites for future virtual screening campaigns (E).
We therefore successfully integrated SeeSAR and infiniSee into a complete hit identification and validation campaign, which allowed us to significantly reduce the biophysical screening effort. The combination of virtual screening and similarity search also reduced the false negative rate in our hands, making both tools valuable programmes within screening campaigns. The use of SeeSAR and infiniSee therefore allowed us to increase the number of compounds screened, ultimately leading to the discovery of novel non-covalent ligands for all Atg8 proteins, demonstrating the druggability of the entire family. A pre-print of the full story is available at the doi: https://doi.org/10.1101/2023.10.05.560930
After 1 year, Martin has achieved the following goals:
- We set our first goal on identification of binders for the LDS of LC3A (A). To achieve this, we used our in-house library of chemical matter consisting of approx. 8500 compounds for virtual screening. We used LC3A as representative member of the Atg8 protein family due to the availability of a co-crystal structure of LC3A and the small-molecule Novobiocin (Ki = 17 µM) (pdb: 6TBE). The native peptide (LIR motif) binds to a hydrophobic binding site which was to our knowledge the only confirmed druggable binding site found within Atg8 proteins. Using this information, we virtually screened all compounds and inspected the binding poses, leading to a selection of 281 compounds which were further validated in fluorescence polarization screening. Subsequently, all positive binders were used for a similarity search in our library yielding 102 novel candidates which were screened in single dose against LC3A, leading to the identification of two non-covalent small molecules with Ki ≤ 10 µM.
- After identification of potential LC3A binders, our second goal was the biophysical characterization of those. We started with KD determination using isothermal titration calorimetry (ITC) for both identified compounds. These measurements showed the expected stoichiometry of 1 and revealed KDs of 10 µM for LY223982 and 1.9 µM for TH152 (B). Therefore, both confirmed small molecules were found to be more potent than any other small molecule found in the literature. Next, we were interested in the family-wide selectivity and found TH152 to be unselective within all Atg8s while LY223982 showed selectivity towards LC3A and LC3B (as known for Novobiocin), making both compounds excellent candidates for tool compounds (C). Last, we utilized NMR spectroscopy to not only measure affinity but also identify the binding site. Mapping all protein-compound interactions to the protein surface successfully revealed the LDS as binding side which is in agreement with the virtually screened cavity (D).
- As a last goal, we were interested in additional binding sites for potential future screening campaigns. We decided to target the LDS due to the two present hydrophobic pockets and the existence of a known peptide ligand which allowed us to set up a biophysical screening system. However, there might be more binding sites present in LC3/GABARAP proteins, which were not identified in the past. To address this, we initiated a structure-based screening using X-ray crystallography together with the DSI-poised fragment library. We soaked 1006 LC3B protein crystals and collected 827 high resolution crystal structures. Refinement of all structures revealed 21 unique fragments bound to LC3B (E). Apart from the known LDS (HP1+HP2) and the UDS, we successfully identified two additional binding sites (S1 and S2) where S1 was found to be highly druggable. Using this knowledge, future virtual screening campaigns might yield additional binders for this so far undrugged binding site.