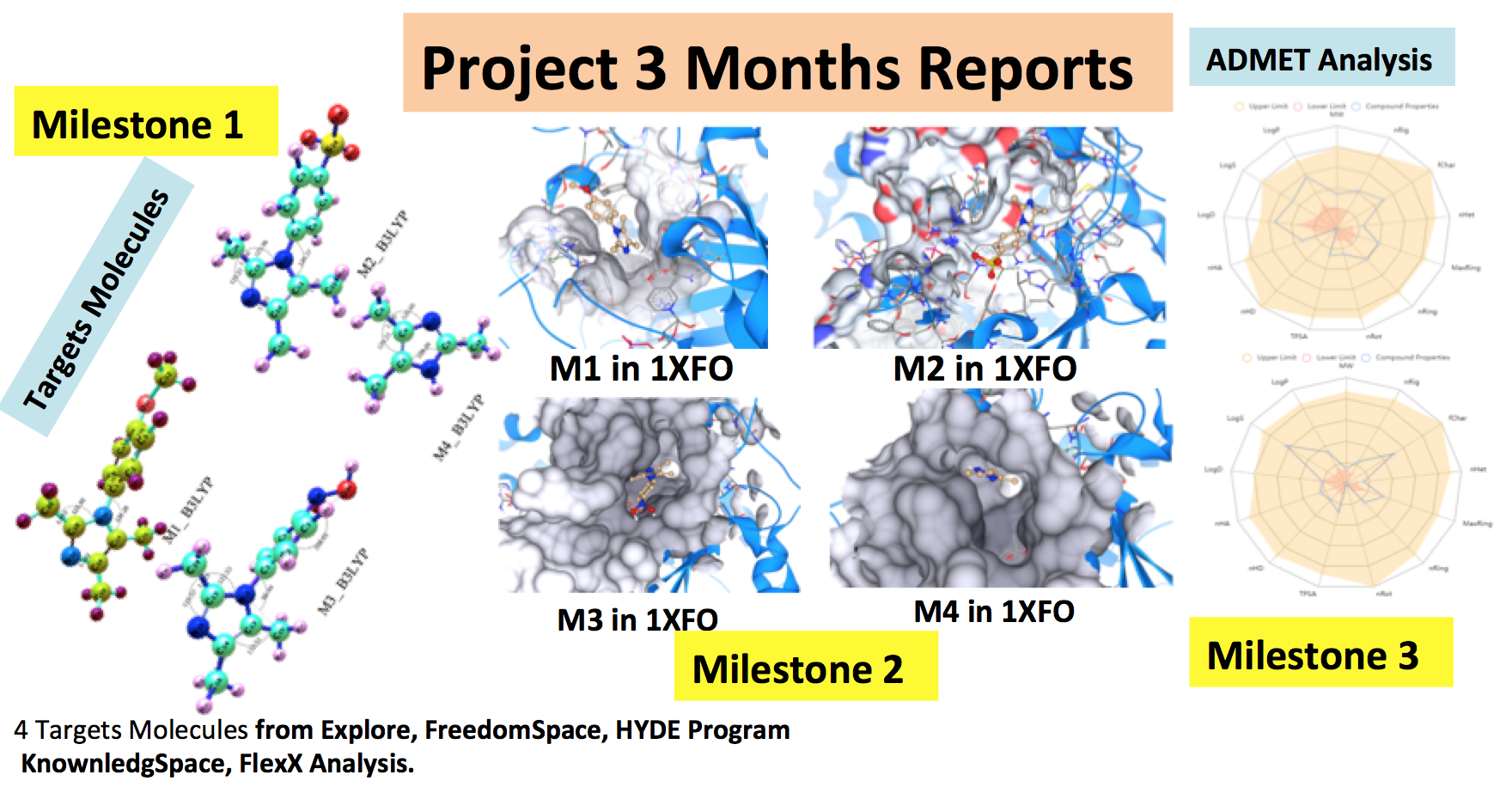
During the first three months, we selected a series of four methyl imidazole derivatives (M1-M4) bio-molecules to treat cancer. Similar chemical and biological properties were obtained by virtual screening using InfiniSee software. To assess the binding interactions between the title molecules and the targeted protein, in-silico molecular studies were carried out. The biological activity values of methyl imidazole derivatives (M1-M4) against various cancer proteins having PDB IDs 3F66, 4XVE, 1XF0, and 5Y8Y were calculated in this work using the molecular docking approach and ADMET analysis. These cancer proteins' biological activities were enhanced by interactions with the methyl imidazole derivatives (M1-M4). The optimized molecules were obtained using Gaussian16W software and saved in PDB format. Molecular docking calculations were done using SeeSAR.12.1.0 software program to screen by bio-simulation, biological activities of title molecules against various cancer proteins.
After 3 months, Simplice has achieved the following milestones:
- The virtual screening planned in this work was successfully carried out on all four series of molecules by Molecular Docking. Biological activities and molecular interactions with target proteins were investigated by ADMET. In infiniSee's scaffold hopper mode, filtering parameters such as LogS, LogP, BBB(Blood-Brain-Barrier), TPSA (Topological Polar Surface Area) enabled us to evaluate Drug-Likeness and ADMET Forecasts and predict Pharmacokinetic properties.
- In SeeSAR, the PDBs of the four target proteins were identified and selected. Interactions at protein sites after molecular cocking were assessed, showing the important sites of the molecules and the types of interactions they might be involved in with residues of the proteins' active sites.
- The crystallographic structures of the four proteins were successfully checking from SeeSAR and the active sites extracted. The ligands complexed with the receptors were then extracted to release the active protein sites. Unoccupied pockets were spotted, and additional residues were then added to saturate the site for automatic site activation, and missing residues were added for improved Molecular Docking and ADMET (Adsorption, Distribution, Metabolism, Excretion and Toxicity). For each target molecules (M1-M4), and by InfiniSee software, Humain Intestinal Absorption (HIA), MDCK Permeability, Caco-2 Permeability, Pgp-Inhibitor, Pgp-Substrate and Plasma Protein of Binding are carried out. Others parameters on Distribution, Metabolism, Excretion and Toxocity are investigated in the same in-silico condition.