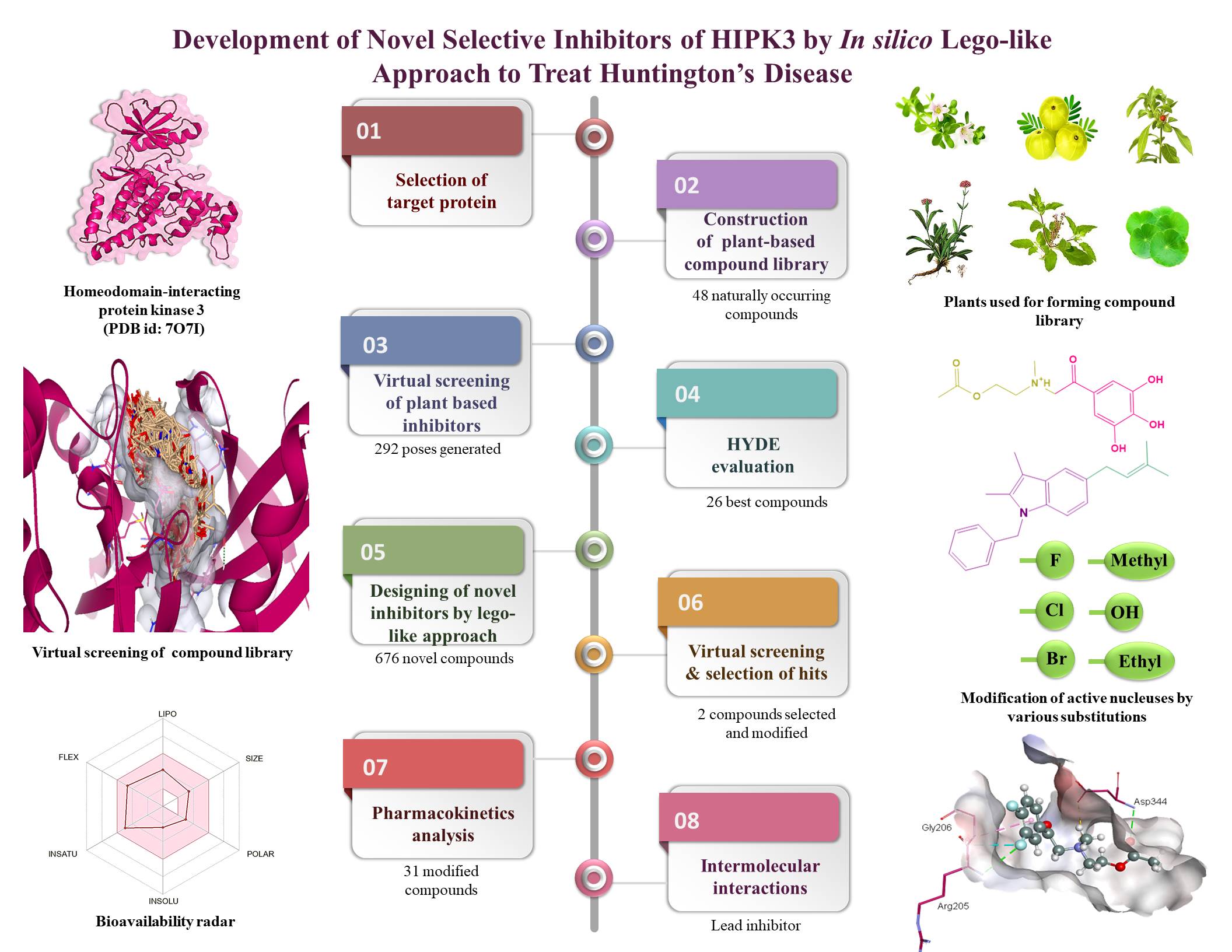
Huntington's disease (HD) stands as a genetic neurodegenerative disorder marked by a cognitive decline attributed to detrimental modifications in the nucleotide sequence of the huntingtin gene (Htt). The aberrant Htt undergoes transcription and translation giving rise to a defected huntingtin protein (mHtt). This mutant variant accumulates within the brain tissues, contributing to the distinctive pathological features of HD. The existing medications merely alleviate the symptoms; therefore, the primary objective of this project was to design highly potent inhibitor specifically to target homeodomain interacting protein kinase 3 (HIPK3). The inhibition or knockdown of HIPK3 has been reported to diminish the mHtt levels in HD patients. Therefore, we employed BioSolveIT tools in our study to examine the protein, identifying binding sites, constructing novel inhibitors by ligating fragments of natural compounds and selecting optimal hits. Our investigation also involved a thorough analysis of the pharmacokinetic properties of each inhibitor and inducing favorable modifications in their structures facilitating the discovery of lead compounds.
After 1 year, Sumera has achieved the following goals:
- Initially, a compound library of 48 biologically active compounds, from six plants having neuroprotective roles, was constructed via PubMed and PubChem. These plants include Withania somnifera, Bacopa monnieri, Centella asiatica, Ocimum sanctum, Nardostachys jatamansi and Emblica officinalis. All these compounds were subjected to energy minimization to improve their binding affinities using ChemDraw 3D. Afterwards, the three-dimensional structure of HIPK3 was selected from RCSB PDB and its most druggable binding site was determined using binding site mode of SeeSAR based on the highest DoGSiteScore. The FlexX functionality of SeeSAR was then used to dock the compounds in compound library to generate poses via standard docking. The docked poses were screened based on their estimated affinities, torsion angles and clashes.
- The structures of the most active natural compounds were analyzed in the analyzer mode of SeeSAR and the fragments (group of atoms) having favorable HYDE were scrutinized. These fragments were combined randomly to create novel compounds utilizing the molecular editor mode of SeeSAR. Throughout the process, considerations such as log P, topological polar surface area (TPSA), and the potential to traverse the blood-brain barrier were integral. All these novel compounds were docked in the most druggable binding site of HIPK3 to generate ten poses for each, which were scrutinized based on highest estimated affinities.
- The ADMET properties of the novel scrutinized compounds were carried out via SeeSAR, SwissADME, ProTox-II and eMolTox. The primary aim of the multisoftware analysis was to reduce the risk of false positive results and to ensure the safety of the synthesized compounds. Ultimately, two compounds were obtained that were non-toxic, non-mutagenic, and non-carcinogenic and followed all the druggable rules. Subsequently, numerous groups were substituted at various places of selected novel compounds to improve their binding affinities via molecular editor mode. One of the two nucleuses (selected novel compounds) showed favorable results with numerous substitutions retaining its non-toxic nature. Therefore, they have been selected and synthesized to further assess their inhibitory potential in in-vitro and in vivo.