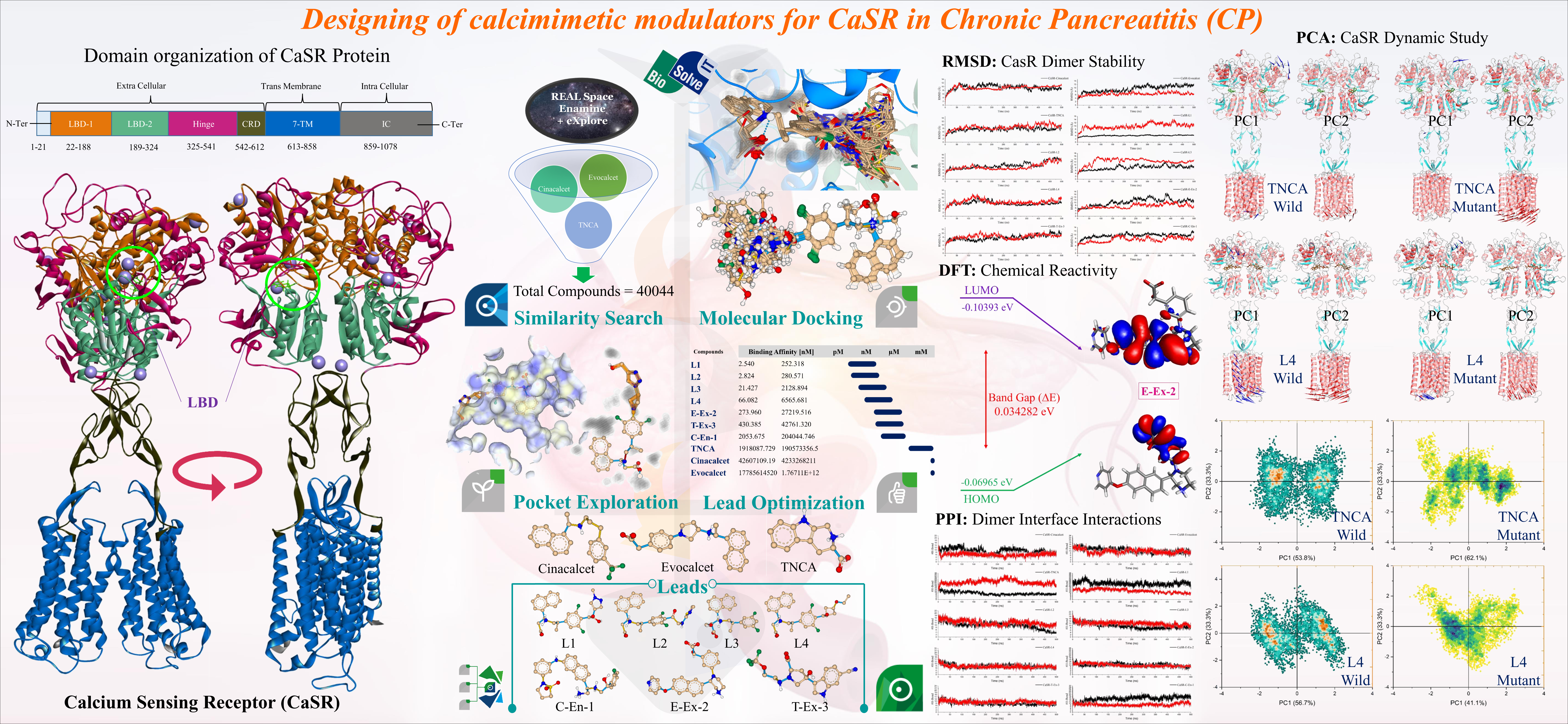
CaSR, a widely expressed Class-C GPCR, detects calcium levels, regulating insulin release, parathyroid hormone, and mineral ion homeostasis. Missense mutations in the CASR gene, such as L173P in the CaSR, are common risk factors for chronic pancreatitis (CP). Calcimimetic molecules (Cinacalcet, Evocalcet, TNCA) show potential for CP. Current CaSR modulators effectively lower PTH but have side effects. For this study, we obtained the active dimer structure of CaSR (PDB ID: 7SIM), conducted mutations, and performed structural preparation and minimization. Using calcimimetic molecules (Cinacalcet, Evocalcet, TNCA), a similarity search produced 40,044 compounds (>95%). Molecular docking with FlexX/SeeSAR for wild-type and mutant CaSR resulted in 30308 compounds (docking scores: -43.48 to 7.69 kcal/mol). Post-screening identified 15113 compounds with better scores than standard molecules, namely Cinacalcet (-12.96 kcal/mol), Evocalcet (-18.39 kcal/mol), and TNCA (-36.31 kcal/mol). Docking showed consistent scores for wild and mutant CaSR, and FlexX reproduced binding poses effectively. We identified 9 hits with the highest docking scores in both chains and optimized them further. Using FastGrow in SeeSAR Inspirator mode, we added favorable functional groups, resulting in 18134 compounds. Simultaneously, ligand pose optimization with Hyde scoring identified a compound with pM binding affinity, leading to the pocket exploration of 1915 compounds. Hyde's scoring highlighted 7 lead compounds with superior binding affinity and docking scores compared to standard molecules. The selected leads exhibited estimated affinities in the nM range, while standard compounds (Cinacalcet, Evocalcet, and TNCA) showed estimated affinities in the mM range. This study affirms that leads from Hyde scoring have a higher binding affinity towards the CaSR receptor, suggesting their potential for modulating CaSR function. Our goal was to restore CaSR mutant activity by maintaining its active dimeric form; therefore, we performed 10 µs of MD simulations on CaSR wild-type and mutant structures, revealing stable complexes formed by selected lead compounds. DFT calculations showed the chemical reactivity of lead compounds is similar to Evocalcet and superior to TNCA and Cinacalcet. This dual approach supports the potential of these leads for modulating CaSR mutant activity. BisolveIT's cutting-edge software suite, including infiniSee, SeeSAR, FlexX, Hyde, etc., empower precise drug discovery in modulating CaSR receptor with unparalleled efficiency, accuracy and reproducibility. Seamless integration facilitates robust exploration, optimization, and analysis, enhancing our understanding of potential therapeutic candidates.
After 1 year, Kiran has achieved the following goals:
- Virtual screening based on similarity search from chemical spaces & natural compound libraries using infiniSee followed by molecular docking in SeeSAR. Employing Enamines REAL space and the eXplore space, we conducted a comprehensive search for structurally similar compounds to Cinacalcet, Evocalcet, and TNCA, resulting in a selection of 40044 compounds. Binding cavities for both wild and mutant CaSR were defined, and molecular docking using FlexX was performed. Our analysis revealed that FlexX consistently reproduced binding poses, as evidenced by the docking of 40044 compounds in replicates within Chain A and Chain B of both wild and mutant CaSR. Docking scores ranged from -43.480 to 7.690 kcal/mol. Following the screening process, 15113 compounds exhibited superior docking scores compared to the standard compounds. We then found common compounds showing the highest docking scores in both chains and were selected as potential hits for further lead optimization.
- Lead optimization of the potential hits retrieved from FlexX docking using FastGrow approach, to fully cover the large CaSR binding pocket. The initial hits were explored using FastGrow in SeeSAR'as Inspirator, generating 18134 compounds. Concurrently, docked poses were optimized with Hyde scoring to predict binding affinities. One compound showed picomolar estimated affinity and was utilized for pocket exploration, generating 1915 additional compounds. From analyzing 50449 compounds with Hyde scoring, we selected 7 lead compounds with estimated affinities in the nM range. These outperformed standard compounds Cinacalcet, Evocalcet, and TNCA with affinities in the mM range. Our findings confirm that selected leads demonstrate significantly higher binding affinities with CaSR receptor. Moreover, these compounds exhibit the potential to modulate CaSR function, marking them as promising candidates for further investigation and development.
- Molecular dynamic simulation and DFT calculations to support the findings. Our objective was to restore CaSR mutant activity by preserving its active dimeric form structure. To achieve this, we conducted 500 ns MD simulations on CaSR wild and mutant type structures, complexed with selected leads and standard compounds. Interface dynamic analysis involved computing intermolecular interactions to assess whether selected lead compounds maintained interactions, thereby retaining or enhancing CaSR dimer stability. MD simulations, along with essential dynamic analyses, confirmed that the selected lead compounds could establish a stable CaSR dimer in presence of L173P mutation. Also, DFT calculations indicated that chemical reactivity of the lead compounds is similar to Evocalcet and superior to TNCA and Cinacalcet. This dual approach provides strong evidence for potential of these leads in modulating CaSR mutant activity and regaining CaSR function in the case of the L173P mutation.