BiosolveIT’s software was so amazing that I felt as if I was on the top of the world with cutting edge tools at my hand during the last one year. I had the pleasure of playing with proteins imported from protein data bank through analysis of protein binding sites, binding pocket active residues, hydrogen bondings, hydrophobic interactions, pi-pi interactions, HyDe for co-crystallized and docked inhibitors. Had tranche of zillions of compounds at my hand in infinisee chemical space navigation tool. Had the delight of construction of custom libraries based on pharmacophore war heads of interest with diverse structures endowed with scaffold hopping abilities. Had the privilege of applying filters for drug-like properties for my compounds of interest and their export as sdf, smiles and exel sheets. Relished the advantage of importing my tailor-made libraries into the drug discovery dashboard SeeSAR. In built pharmacophore generation tools were super easy to understand and user friendly. One click Docking with estimated affinity calculations, torsions, inter and intramolecular clashes, labelling of residues, topological surface view, focus view, screen shot generation. Hyde scoring halos provided me with bird’s eye view of favorable and unfavorable binding affinities at a glance. Had the advantage of marking my ligands of interest not only as active or inactive but also liked the tool for adding annotations to any molecule. Enjoyed the freedom of construction of “absorption, distribution, metabolism and excretion” (ADME) property plots. Had the benefit of playing with inhibitors of interest in inspirator mode for inspection of any useful modifications that may be done. Because of these tools I was able to achieve my dream goal of finding common molecules that hopped and docked successfully on different scaffolds of four proteins of my interest having roles in cancer spread.
After 1 year, Dr. Jariya has achieved the following goals:
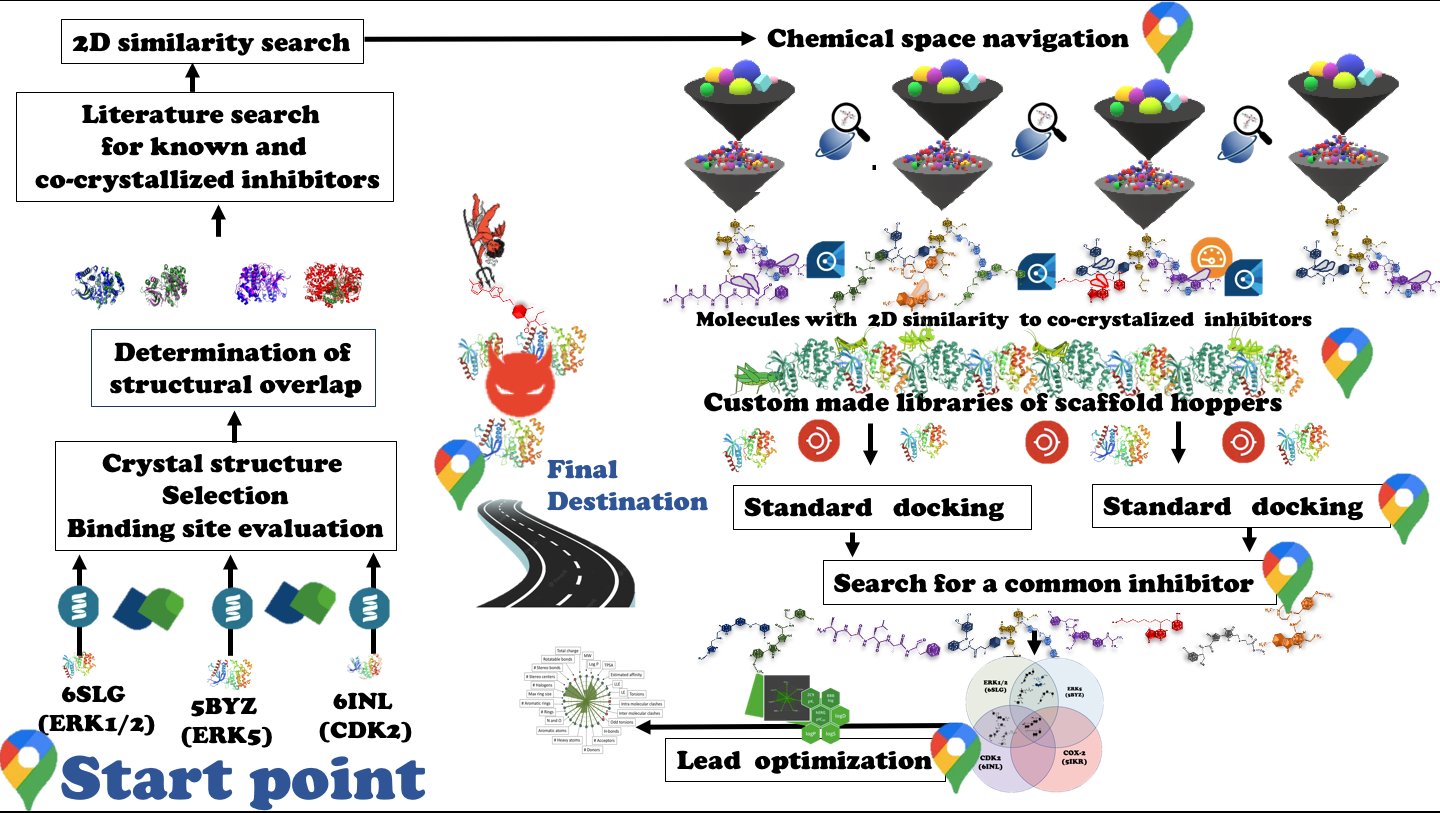
- Analysis of protein binding sites, construction of custom-made libraries of chemicals, docking. Crystal structures of kinases involved in cancer spread were analyzed. Their binding sites were ear marked. Their documented inhibitors were subjected to 2D similarity search with their pharmacophores as reference points to navigate the chemical spaces. The search parameters were tweaked to 100 results per space with similarity scale of 0.7 to 0.9. A cache of diverse compounds was retrieved, with abilities of scaffold hopping due to possession of similar pharmacophore war heads. The retrieved molecules were then docked against these kinases. Protein-molecule complexes were then filtered for estimated affinities less than 500nm. The shortlisted complexes were then compared to find common inhibitors. Few potential common inhibitors were located. These were thoroughly analyzed for their interactions with binding sites and their ADME properties. Their pose views and property plots were generated
- The results obtained by SeeSAR were validated by docking of same molecules with Autodock vina. The crystal structures of enzymes involved in cancer spread were aligned with help of pymol to see their evolutionary commonalities. Three out of four enzymes showed favorable alignment with root mean square deviation RMSD less than 2. However, one enzyme failed to align with favorable RMSD. But, the proposed inhibitors were able to bind it because of their scaffold hopping abilities.
- Currently I am doing molecular dynamic simulations for proposed inhibitors. I have also contacted vendors for in-vitro testing of proposed inhibitors. However, WuXi lab network has given quote of 3230$ for 10 grams each of two proposed inhibitors, which is far far beyond my buying power. And Chem space team has given quote of 2198 Euros which is also not my buying power. Otherwise I could have performed invitro testing of these chemicals as well