Peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors playing a critical role in the regulation of metabolic homeostasis, and thus represent valuable therapeutic targets for the treatment of metabolic disorders.
The PPAR family comprises three different subtypes: α, β/δ, and γ, whose expression and actions differ according to subtype, organ, and tissue cell type.
The development of more balanced drugs interacting with PPARs, devoid of the side-effects showed by some marketed agonists, is considered a major challenge.
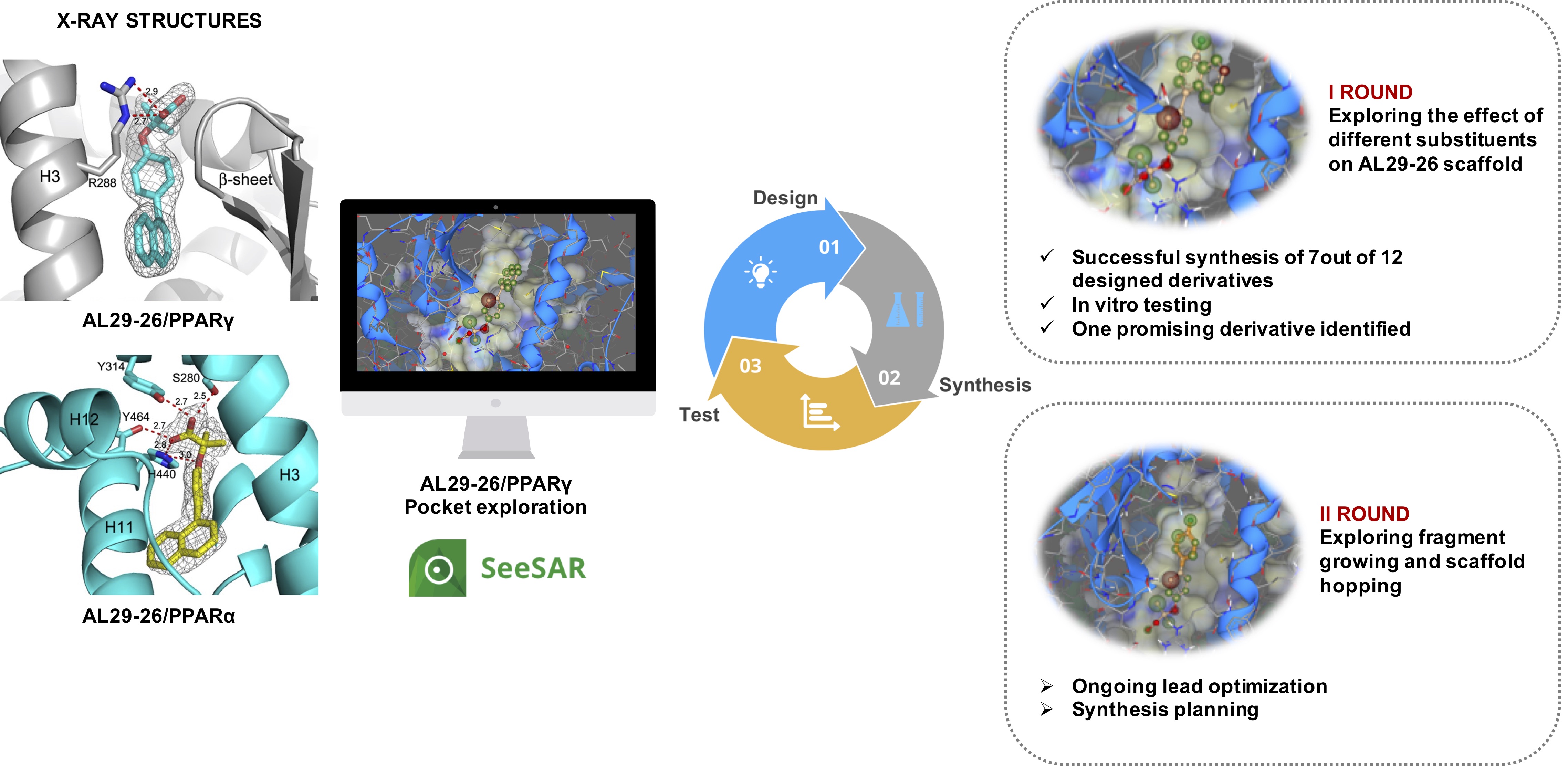
We recently reported a novel PPAR pan-agonist, AL29-26, with an attractive and balanced activity profile, which represents an interesting lead of a new class of drugs for treatment of metabolic diseases and type 2 diabetes. Moreover, we solved the crystal structure of AL29-26 in complex with PPARα and PPARγ. Therefore, we embarked on a lead optimization campaign focused on the lead compound AL29-26 to improve its potency and activity in cell-based assays.
Our strategy involves iterative cycles of design, synthesis and biological tests of the hypotheses generated in silico with the support of SeeSAR protocols, kindly provided by BioSolveIT. The user-friendly SeeSAR platform was used to carefully analyze the crystal structure of AL29-26 in complex with PPARγ. Such analysis allowed to carry out the structure-based design of AL29-26 close analogues, based on the ligand crystallographic pose. In the first round of hypotheses design, we inserted different small substituents on AL29-26 scaffold, with the help of Molecule Editor, Inspirator, as well as affinity estimations calculated by HYDE. We quickly designed 12 analogues, which were proposed for synthesis. Out of the 12 designed analogues, 7 were eventually synthesized while 5 were discarded because of issues during synthesis work up. A coactivator recruitment assay was then set up, measuring the ability of each compound to stimulate the interaction of PPARγ with a coactivator peptide SRC-1. Among the synthesized analogues, one compound, derivative AL29-26-3, emerged as the most promising one. Additional studies, including further cycles of design, with the help of SeeSAR, and synthesis planning are currently ongoing. The obtained compounds will be tested again using the above-described coactivator recruitment assay as well as in transactivation assays on HepG2 cells; moreover, their effect on the expression of selected PPAR target genes involved in lipid accumulation and insulin sensitivity, will be evaluated by real-time quantitative PCR (RTqPCR).
After 1 year, Carmen has achieved the following goals:
- Design and synthesis of AL29-26 derivatives. As an initial step, we performed a careful analysis of the crystal structure of AL29-26 in complex with PPARγ. The SeeSAR platform allowed to easily explore the ligand binding domain of the receptor, whereas the tools Molecule Editor and Inspirator were used to insert different substituents on the molecular scaffold of AL29-26. The effect of different substituents was analyzed on the fly by means of HYDE. We prioritized compounds with favorable estimated affinity, lacking molecular clashes with protein residues. Appropriate synthetic routes were devised, which afforded 7 out of 12 designed compounds.
- In vitro testing of the synthesized AL29-26 derivatives. The activation of PPARγ was measured by Alpha Screen Technology in a coactivator recruitment assay, in order to measure the agonistic activity of the designed compounds. Among the tested compounds, derivative 3 (AL29-26-3) displayed the highest efficacy. Again, SeeSAR was helpful in analyzing the structural determinants responsible of receptor modulation by the synthesized derivatives.
- Second round of AL29-26 optimization. The obtained results prompted a further cycle of hypotheses design to be proposed for synthesis. The SeeSAR platform, together with the built-in fragment library was proficiently used to design further derivatives. Scaffold hopping opportunities were also explored, by taking advantage of the ReCore tool. The designed derivatives are currently being analyzed for synthetic feasibility. Besides the above-described coactivator recruitment assay, we plan to test the synthesized compounds in transactivation assays on HepG2 cells. Moreover, for the most active ones, their effects on the expression of selected PPAR target genes involved in lipid accumulation and insulin sensitivity, will be extensively evaluated by real-time quantitative PCR (RTqPCR).