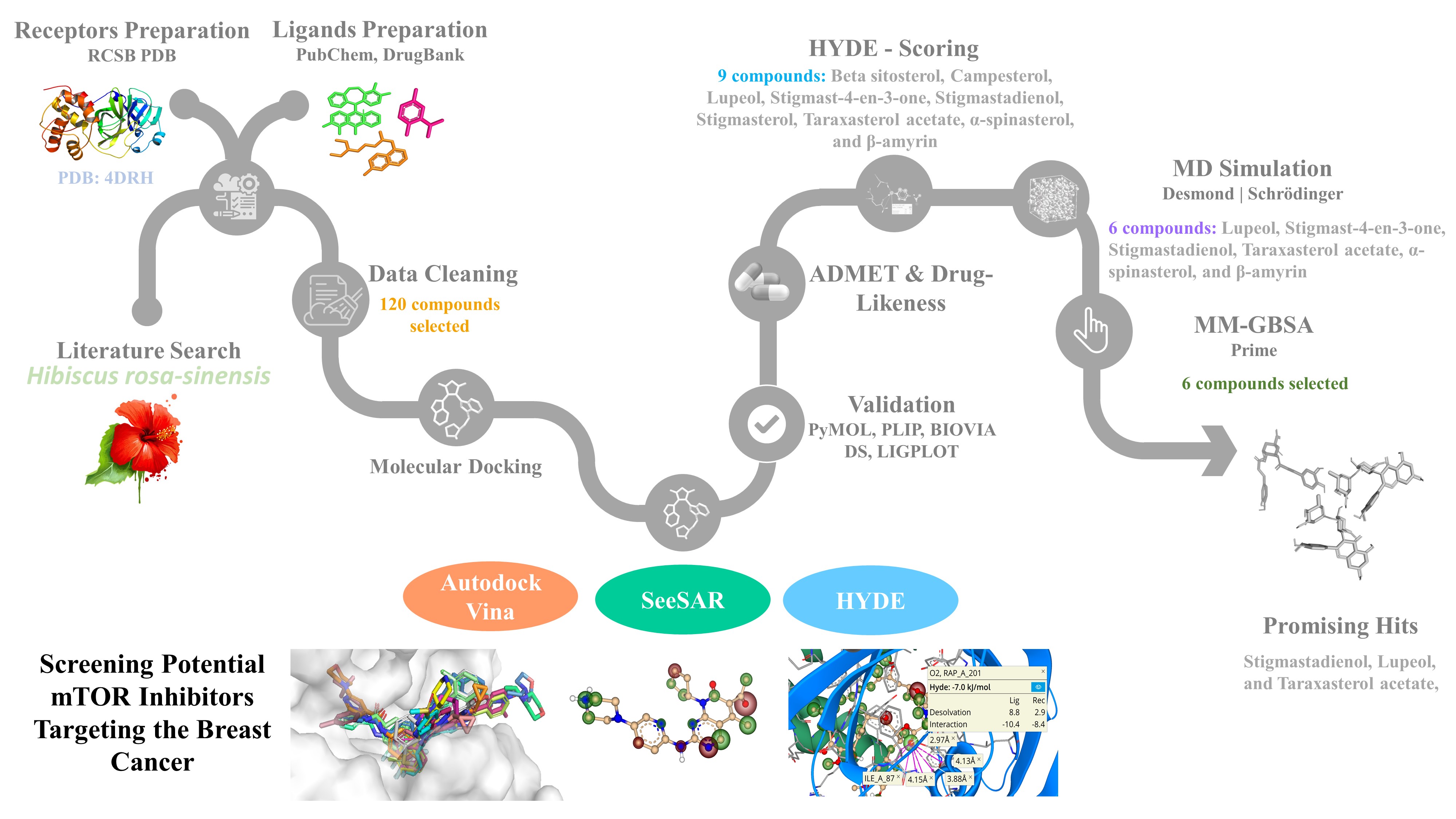
-Aapproximately 120 Hibiscus rosa-sinensis components and ten approved anti-cancer drugs, including the mTOR inhibitor everolimus, were used in the comparative analysis. With the help of SeeSAR, we were able to locate the druggable pocket in the mTOR (PDB ID:4DRH) protein. In addition, the druggable pocket was determined, and the empty spaces were examined in detail. The compounds with the highest ranked binding poses were loaded using the SeeSAR tool and the HYDE scoring to give interactive, desolvation, and visual ΔG estimation for ligand binding affinity assessment. We are employing two distinct strategies for the molecular docking of the discovered ligands. First, Autodock Vina was used to docking the compounds to mTOR. Second, the molecules that were chosen were "docked" with mTOR to see how well they worked in SeeSAR. As a result, 16 molecules with the best affinity were selected for further analysis. Following that, several poses of selected ligands were evaluated using HYDE calculations, and 9 ligands were chosen. By using Lipinski's rule of five to the chosen compounds, the ADMET profile and drug-likeness characteristics were further examined to assess the anti-breast cancer activity. Following, the prospective candidates underwent three replicas of 100 ns long molecular dynamics simulations, preceded with MM-GBSA binding free energy calculation. The stability of the protein-ligand complex was determined using RMSD, RMSF, and protein-ligand interactions. The results demonstrated that the best mTOR binding affinities were found for stigmastadienol lupeol, and taraxasterol acetate, which all performed well in comparison to the control compounds. Thus, bioactive compounds isolated from Hibiscus could serve as lead molecules for the creation of potent and effective mTOR inhibitors for breast cancer therapy. Finally, this study has been published in Molecular Diversity Journal with the DOI: https://doi.org/10.1007/s11030-022-10556-9
-SeeSAR and HYDE were utilized to load the compounds with the highest-ranked binding poses of Vina into HYDE for visual affinity assessment post-scoring. The compounds were created using the SeeSAR-integrated editor. SeeSAR visualizes the expected free energy of binding (ΔG) using "coronas" ranging from dark red (extremely unfavourable for affinity) to gigantic spherical dark green (extremely favourable). The tool provides for the interactive examination of energy and torsions (measured in kJ/mol), the enthalpic (interaction, ΔH) and the desolvation (dehydration, −TΔS) contributions to protein and ligand binding. Additionally, SeeSAR allows for a semi-quantitative calculation of the thermodynamic profile for all chemicals studied.
After 1 year, Hezha has achieved the following goals:
- Performing structure-based virtual screening via Vina and SeeSAR. The initial goal of this work was to screen a number of inhibitors derived from Hibiscus rosa-sinensis for the possible targets to inhibit the mTOR and to determine which has the greatest affinity for the receptor. There has been a comprehensive review of the literature. The crystalline structure of the PPIase of FKBP51 with rapamycin co-crystallize was employed to provide light on the binding mode of the selected medicines to mTOR. About 120 different Hibiscus rosa-sinensis components were tested for their ability to bind to a breast cancer protein target, and the results were compared. Ten FDA-approved anti-cancer medicines, including everolimus, were employed as the positive controls. 16 molecules with the best affinity were selected for further analysis. These docking complexes were checked for lipophilic ligand efficiency, torsion quality, intra and inter molecular clashes.
- Binding assessment visualization based on desolvation and interactions. SeeSAR, which proved to be user-friendly software and provided data that were simple to read, was used to continue the investigation as our second goal. The docked poses of a few molecules were examined using SeeSAR and HYDE scoring, showing crucial, unique, and interpretable conformations. At first, all mTOR protein binding sites were investigated. The interaction between the ligand-protein complexes was then observed using the reference ligand everolimus and its binding pocket, and the results were promising. PyMOL, PLIP, BIOVIA DS, and LIGPLOT were used to investigate the interactions of targeted protein active pocket and rapamycin. Later, a HYDE estimation-based evaluation of various poses of chosen ligands was carried out.
- ADMET properties, molecular dynamics (MD) simulations and MM-GBSA Calculations. Another aim was the investigation of drug-likeness and in-silico ADMET calculation utilizing integrated online platforms ADMETlab and Molinspiration. This is because all potential medication candidates must demonstrate the strong biological activity with low toxicity. The development of a successful novel medication candidate relies heavily on early pharmacokinetic evaluation. The most active compounds and two FDA-approved drugs were subjected to the calculation. Subsequently, the Schrodinger/ Desmond software was used to run MD simulations on the selected docked complexes at 100 ns time intervals to examine receptor stability and ligand interaction. In addition, the relative binding free energy for each mTOR-ligand combination and the reference complexes was computed using the molecular mechanics generalized Born surface area (MM/ GBSA) calculation.