The Tumour microenvironment (TME) is becoming a significant challenge to the existing chemotherapies, and it was found to be associated with the metabolic reprogramming in the cancer cells. As per the Reverse Warburg effect, the key regulators of the lactate shuttle, monocarboxylate transporter 1 and 4, plays a significant role in the evolution of TME, which is responsible for favouring tumorigenesis. This work aimed to design an MCT inhibitor to target the tumour microenvironment, and the work plan extended to experimenting with the potential of these designed MCT inhibitors as a chemotherapeutic or cytocidal moiety. In short, this work aimed to design an effective molecule to target both the tumour cells and their microenvironment with a single moiety.
Initially, a ligand library was generated by an extensive search in various databases, where all of them could serve as hits for developing a new inhibitor for the MCT-4 receptor. They were checked for affinity towards the target protein via docking with the top hits. These selected lead molecules were exported to inifinisee, and molecules with a similarity of 9 and above have opted for conjugation with chemotherapeutic agents. The conjugated molecules were re-docked and modified by replacing the various elements of the lead molecules and by filling the unoccupied space in the binding pocket. Their properties also were calculated to choose the ligand-like molecules. Thus, some promising molecules arose to inhibit the growth of cancer cells by silencing the tumour micro-environment and targeting tumour cells.
A valid 3D-QSAR model and a best pharmacophore hypothesis for the cancer receptor were generated and statistically validated in the next step. These models predicted the activity value for those designed molecules against the cancer receptor. The molecules with the highest predicted activity value and the best-fit score for the top-generated pharmacophore hypothesis were selected for a multi-targeting approach. These molecules were docked for their binding affinity against the cancer receptor and screened for their amino acid interactions with the binding pocket of the target protein. The top hits were further analyzed for the stability of the protein-ligand complex through molecular dynamics (MD) studies after the ADME analysis and toxicity predictions. The best molecules can be proposed as promising candidates to target the tumour cells and microenvironment.
I am so thankful to the BioSolveIT scientific challenge for this excellent opportunity to be a part of this scientific challenge and for providing me with the SeeSAR and infiniSee software, which played a crucial role in the designing strategy and other computational tools explored in the final QSAR and MD protocols.
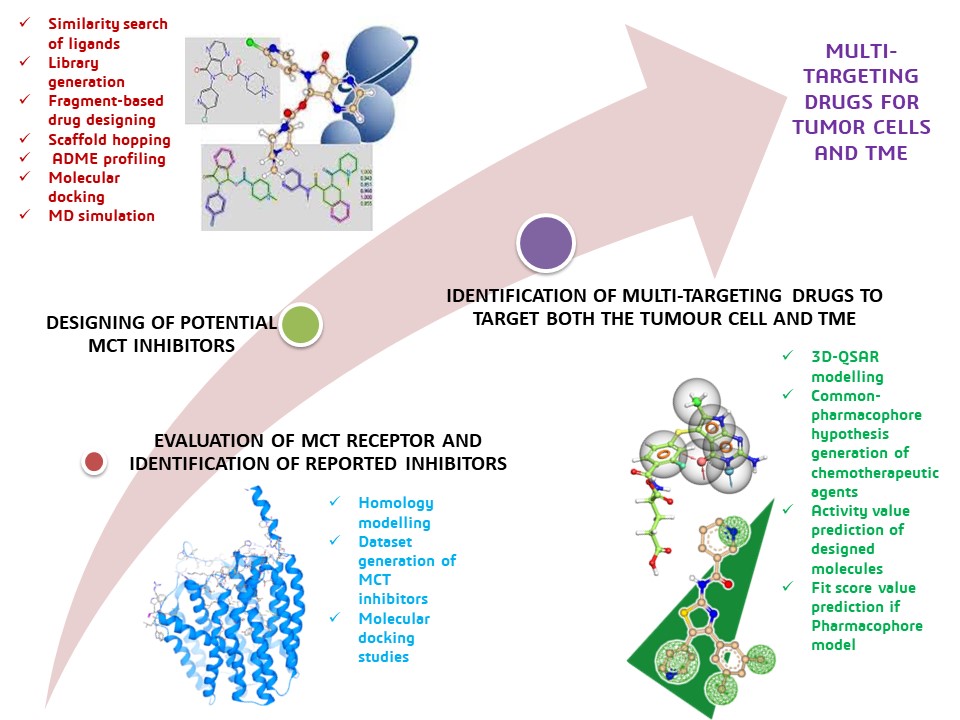
After 1 year, Sonu has achieved the following goals:
- Evaluation of MCT receptor and identification of its inhibitors: An initial library was created by searching for all the possible reported compounds against all the Monocarboxylate transporter protein family members. Various databases such as PubChem, ChEMBL, DRUG BANK, ZINC-15, BINDINGDB, ChemSpider and IUPHAR/ BPS Guide to Pharmacology were extensively searched and all the possible ligands from various pieces of literature, clinical trials, patents, and all the likely reported works were extracted. An initial library of about 3855 molecules was generated and validated for their properties. They were subjected to basic filtering via docking mode, which started with the preparation of homology modelled and validated 3D-PDB structure of MCT-4. All the possible binding pockets were analysed, and the best one was selected for docking studies. The top hits were screened through Virtual Hit Triage based on their expected affinity and other features.
- Designing of a potential MCT inhibitor: The top hits were selected as the lead molecules and exported to the InfiniSee to generate diversified structures based on the lead molecules that show the highest affinity towards the target protein structure. All the possible compounds were generated as an extensive library, and the compounds with the highest similarity of 0.9 and above were considered the best ligand-like molecules for MCT inhibition. A fragment-based drug designing and scaffold hopping approaches were employed to generate a set of promising molecules. The molecules were then subjected to molecular docking against the MCT receptor to check their binding affinity towards the receptor, and ADME profiling was used to evaluate their molecular properties. MD simulation was also performed to identify the stable drug-protein complexes with which an efficient therapeutic outcome can be anticipated.
- Identification of multi-targeting drugs to target both the tumour cell and TME: A particular cancer receptor was selected, and its reported inhibitors with biological activity value were collected as a dataset. It was then utilised to give rise to a valid 3D-QSAR and common-pharmacophore model. The best models were identified with the statistical evaluation. With the best QSAR model, the activity value of the designed molecules against the particular cancer receptor was predicted, and the top-selected common pharmacophore hypothesis was employed to identify the best-fit molecules. The top hits were selected and subjected to molecular docking studies followed by dynamic simulations. The top hits with higher affinity towards the cancer receptor were screened for their docking scores, and the stable protein-ligand complexes were identified via MD studies. Thus, it resulted in a series of multi-targeting and promising candidates with which we can target both the Cancer cell and their TME.