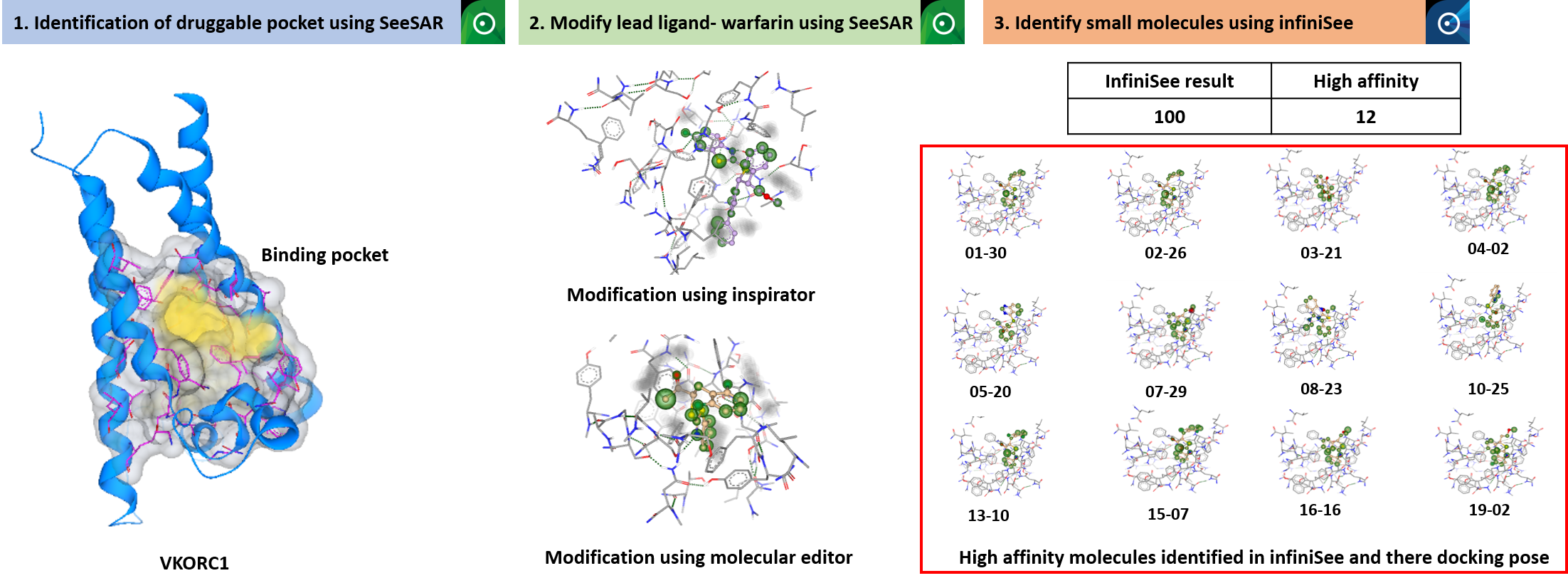
We have identified one druggable pocket in VKORC1 protein using SeeSAR. Further, this druggable pocket was defined and empty spaces were analyzed. Warfarin (the lead ligand) was modified to obtain similar molecules which will not bind with the warfarin resistant residues known in VKORC1. We are taking two different approach for modifying the lead ligand. First, we are modifying it in SeeSAR using molecular editor and inspirator mode. These modified molecules were then dock with VKORC1 to check affinity. Second, we have searched ligands in infiniSee platform by taking warfarin as a template and further docked these molecules to obtain complexes with best affinity. By doing so, we obtained 12 molecules out of 100 with high affinity and lipophilic ligand efficiency. We will further modify the warfarin further and also the 12 new small molecules that showed higher affinity, to obtain a ligand that is not binding to warfarin resistant mutants.
After 3 months, Suvoshree has achieved the following milestones:
- The VKORC1 protein was extracted by removing the tag and adding the missing residues to the known crystal structure template of VKORC1 (6wv3) obtained from PDB. The extracted protein was uploaded in SeeSAR in protein mode (without extracting the ligand warfarin), followed by the analysis in the binding site (show unoccupied pockets). This search lead to the identification of only one druggable pocket comprised of 38 interacting residues (Gly21, Leu24, Ser25, Ala28, Leu29, Lys32, Val56, Phe57, Ser59, Trp61, Gly62, Phe65, Leu67, Ser81, Asn82, Ser83, Phe89, Tyr90, Leu92, Ser111, Val114, Ser115, Ala117, Gly118, Ser119, Tyr121, Leu122, Ile125, Leu126, Leu130, Cys134, Val136, Cys137, Thr140, Tyr141, Asn144). This pocket has similar binding residues as the warfarin binding site.
- The binding site of warfarin in VKORC1 was identified that showed 30 binding residues (Gly21, leu24, Ser25, Ala28, Leu29, Lys32, Phe57, Gly62, Gly64, Phe65, Leu67, Val68, Leu78, Ser81, Asn82, Ser83, Phe85, Gly86, Phe89, Tyr90, Leu92, Ser111, Val114, Ser115, Ala117, Gly118, Ser119, Tyr121, Thr140, Asn144). The unoccupied space was analyzed under visualization tab. Then warfarin was modified using molecular editor and inspiratory mode in SeeSAR platform.
- In parallel to modification of the lead ligand, small molecules were searched in the infinisee platform (REAL Space, CHEMriya, GalaXi, and KnowledgeSpace) by taking warfarin as a template which showed 100 similar small molecules. Then standard docking was performed with these molecules on VKORC1. Out of 100, 12 molecules with best affinity were selected for further analysis. These docking complexes were checked for lipophilic ligand efficiency, torsion quality, intra and inter molecular clashes. Further these 12 molecules will be modified in order to generate a ligand which is not interacting with warfarin resistant residues.