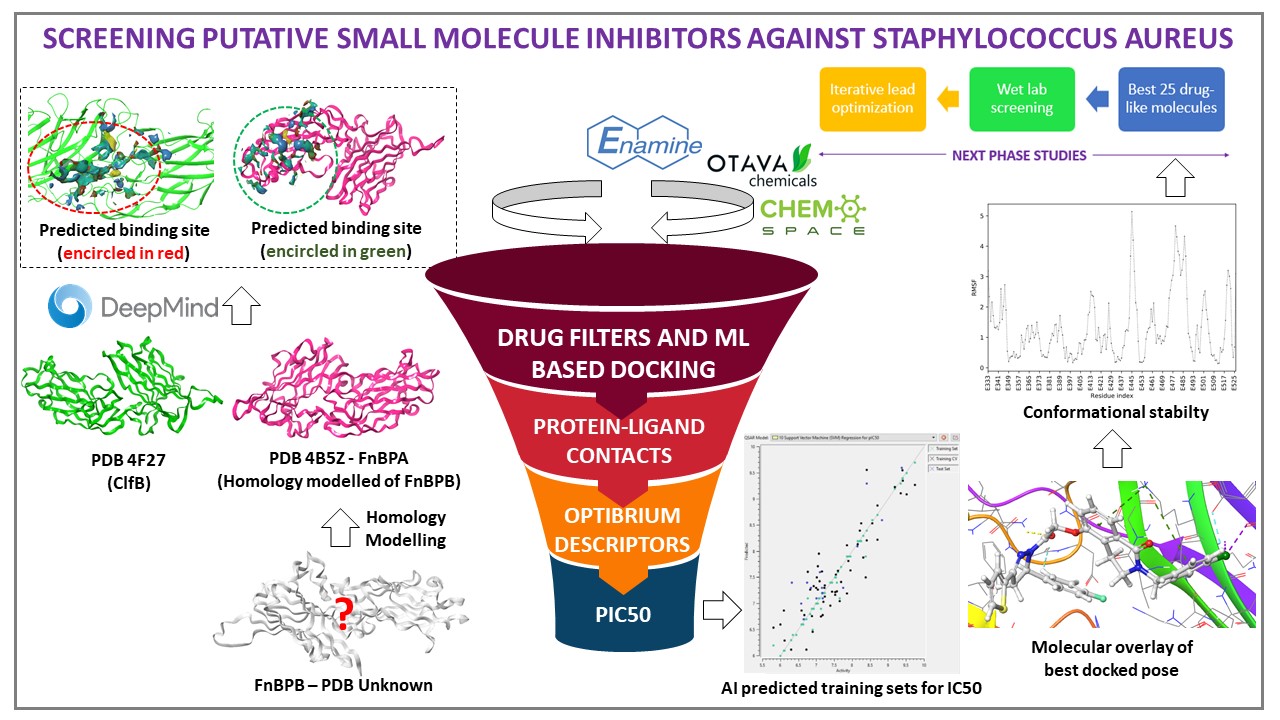
Fibronectin binding protein B (FnBPB) was homology modelled and structure was validated by SWISS MODEL. The drug targets Clumping factor (ClfB) and Fibronectin B binding protein (FnBPB) were subjected to site prediction using open source CASTp software and SiteMap. ClfB was bound to peptides which were huge compared to small molecules. We accurately predicted the probable binding sites for small molecules based on druggability score, surface volume and literature. Then, we did molecular probings (~5K) against predicted sites using infiniSee integrated databases. Violations of Lipinski rules were avoided using DataWarrior. IC50 was predicted using AI based regression and classification model of Scikit-Learn. Based on SeeSAR's descriptors (LE, LLE, torsional class strain and HYDE binding estimations) and ADMET profiling ( Optibrium Properties, PKTools 3.0 and ADMETlab 2.0), top 25 molecules were subjected to MD simulations (conformational stability). Drug synthesis and assay development are underway.
After 3 months, Smriti has achieved the following milestones:
- ClfB (PDB 4F27) was found out from Uniprot and homology modelled proteins FnBPB (PDB 4B5Z – A0A0H2XKG3 was retrieved from SWISS MODEL expasy. Structure of both proteins were validated by comparing structures available with DeepMind – AlphaFold database. These models were accurate (>90% confidence levels). The retrieved PDBs were prepared and optimized. pI was calculated using http://isoelectric.org/ and found to be 5.18. Both proteins were in apo form, and active binding sites for small molecules were not known. CASTp and SiteMap predicted binding sites of both proteins and a validation was done based on reported literature. The binding site lied between N2-N3 region and Fibronectin binding repeating subunits for ClfB and FnBPB, respectively. Further, sites were scrutinized based for druggability score and surface volume. Druggable site was further defined with the help of Atomic Sequence Language tool where residues in 4Å proximity residues were selected for grid generation.
- The anti-bacterial libraries were designed based on commercial availability and synthetic feasibility from Enamine and OTAVAChemicals. These libraries were refined based on the appropriate drug filters with avoidance of violations using DataWarrior. All these refined molecules were probed against the best druggable predicted sites of both the proteins using Xtra precision (Glide) and validated using SeeSAR 11.0. The generated hits were further analyzed for their estimated binding affinities (HYDE), hydrogen interactions, LE, LLE and torsional clash strains and Optibrium descriptors. Simultaneously, the same database was on to do AI based prediction of pIC50 using regression and classification analysis of Scikit-Learn to find the best drug-like molecules. The physicochemical and ADMET descriptors were studied using PKTools 3.0 and ADMETlab 2.0. The top 25 molecules with estimated affinity range at nM-uM range were subjected to further Molecular dynamics simulations.
- The top lead hits are to be procured from the concerned in-stock commercial vendors. Few synthetic feasible molecules will be synthesized with the support of synthetic complexity score (SCScore), using https://askcos.mit.edu, and Spaya AI (https://spaya.ai/) score. The assay to validate inhibitory activity is to be designed further in upcoming months. The procured compounds will be subjected for wet-lab screening analysis. Based on the wet-lab screening results, iterative medicinal chemistry optimizations and point of randomizations will be performed for further improvement in inhibitory activity and ADMET using the Molecular Inspirator and ReCore module of SeeSAR. We are also planning to increase our library chemical space using fingerprint similarities search from the infiniSee chemical space searches based on the promising dual inhibitor scaffold from the screening results. This will allow us for improvement in designing new drug-like molecules.