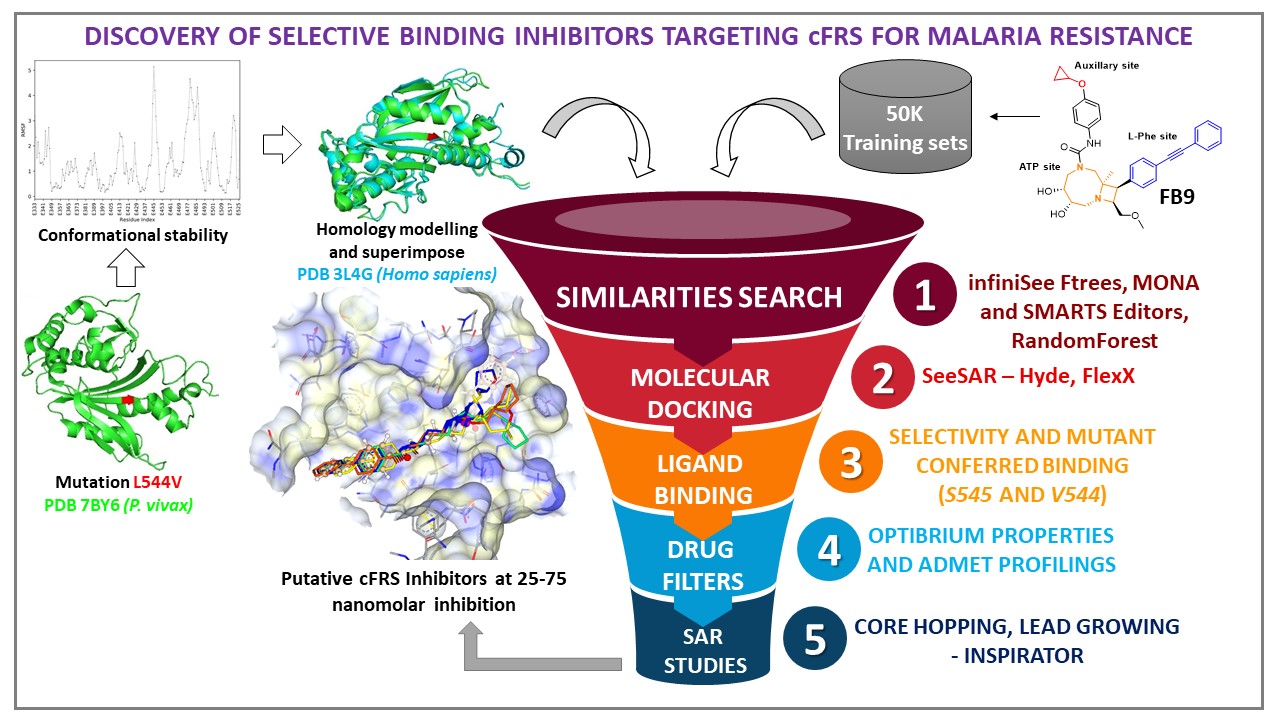
Malaria is caused by apicomplexan parasites, has proven to be the deadliest infectious disease. It is an overwhelming global burden in the tropical and sub-tropical regions of the world. The treatment of malaria is becoming more challenging due to emerging drug resistance from traditional antimalarial drugs and relapses due to various plasmodium strains. To overcome this drug resistance, validation of new drug targets and drug molecules is much needed. Ideally, a new target possesses a new mechanism of action, expedites reduction of the parasite in the erythrocyte stage, and retains efficacy in all stages of the life cycle of the parasite in the human host. The cytosolic Phenylalanine tRNA Synthetase (cFRS) is indispensable for the synthesis of proteins of the parasite. Inhibiting cFRS leads to cell growth disruption. Probing scaffolds selectively binding will result in novel inhibitors which will be taken to the next phase of the drug discovery pipeline. Currently, the reported inhibitor is weakly/moderately potent possessing molecular obesity. We predominantly focused on the mutations in cytosolic tRNA synthetase of Plasmodium vivax conferring drug resistance. Initially, we created the mutation (L544V) in P. vivax cytosolic phenylalanine tRNA synthetase (cFRS) that causes drug resistance. We superimposed the structures of mutated P. vivax (cFRS) with Homo sapiens (HsFRS) to study cytotoxicity effects. This mutated PDB of P. vivax was superimposed onto HsFRS PDB to study novel scaffolds as the inhibitors. In this study, we applied receptor-guided drug design tools from BioSolveIT including FTrees, infiniSee, SeeSAR, and analysis tools such as MONA and SMARTS Editor. From the initial virtual screening, a refinement was done for pose conformers which were focused mainly at the active sites (L-Phe, ATP, and auxiliary), L544V (mutant), S545 (active binding for plasmodium inhibitory), and other active hydrophobic residues. Lead optimization and binding affinity studies were performed for the improvement in binding conformers. The best conformers were further scrutinized as per the Optibrium and ADMET profiling. Based on the synthetic accessibility, synthesis and screening led us to identify ten putative cFRS inhibitors selectively binding at nanomolar concentration range. Further assessment with the support of translational research studies will yield a clinical drug candidate.
After 1 year, Jeevan has achieved the following goals:
- Initially, we have developed a mutated model (L554V) of cFRS from the Plasmodium vivax protein PDB 7BY6 (residue L554V). The mutated model was confirmed with its conformational flexibility (RMSF) and found to be stable without any restraints. The confirmed model was further superimposed on HsFRS (PDB 3L4G) for studying its cytotoxicity effect. Based on a similarity search with the template molecule (FB9), a virtual library was designed (50000) using FTrees and infiniSee. The resultin molecules were further screened using Random Forest and RDKit Fingerprints yielding 2600 molecules. The top 5% of molecules based on the binding affinity determined by SeeSAR and further iterations. This permitted us to yield an effective exploration of scaffold hopping from huge chemical space that could be the target-relevant in a highly time-efficient manner. The best diverse set of 500 molecules with specific structural features and patterns were refined using MONA and SMARTS Editor drug filters.
- The SeeSAR tool helped us to gain a deeper understanding of the energetics of structural features of the molecules that are needed for the selectivity Pv cFRS inhibition. Drug molecules exhibiting favorable potency and efficacy were identified through an evolutionary screening methodology. The best diverse set of series (500 molecules) was probed against the defined binding site of the superimposed drug targets. Few best leads had a low binding affinity range but could not fit into the binding pocket and had poor Optibrium properties as well. The top fifty leads were optimized using the empirical point of randomization and structural modifications using Inspirator to curtail these pitfalls. The newly optimized twenty molecules possess better binding affinities (picomolar to the nanomolar range), key binding residue contact (S545), and optimized Optibrium descriptors.
- All twenty molecules were retro-synthesized to successfully synthesize at milligram levels. The wet-lab screening for functional studies, enzymatic activity, selective inhibition, and cytotoxicity effect were performed as per the well-established protocols by our research group. From the synthesized series, ten molecules promisingly exhibited plasmodium selectivity inhibition at 25 – 75 nanomolar range in triplicates. However, due to the time constraints imposed by COVID-19, and partial functional of wet-lab screening activities no further biological assays were able to be undertaken in this timeframe. Currently, we are screening other molecules with available plasmodium strains (CQ sensitive and resistant) for further assessment for translational research to yield with the clinical drug candidates. The identified drug molecules will support us understand the binding modes and further designing the derivatives with the support of CoLibri.