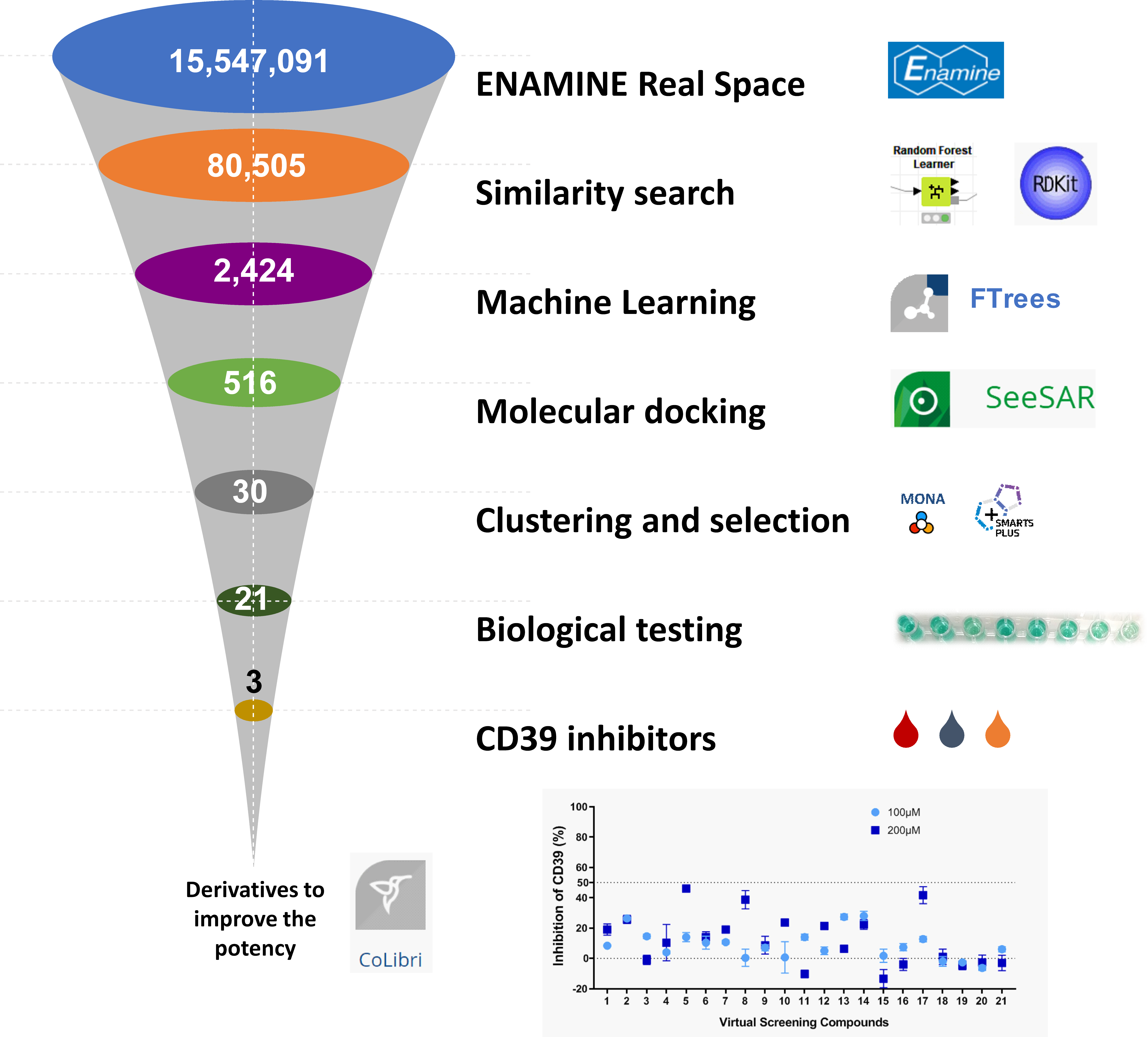
CD39 and other ectonucleotidases control extracellular nucleotide and nucleoside levels, especially those of ATP and adenosine. Inhibitors of CD39 has the potential checkpoint for the immunotherapy of cancer and infection. Currently, the reported inhibitors are only nucleotide analogs and polyoxometalates which are weakly/moderately potent. In this study, we applied ligand- and structure-based drug design tools from BioSolveIT includes FTrees, SeeSAR and analysis tools such as MONA and SMARTS Editor to identify novel, potent and selective non-nucleotide CD39 inhibitors. Screening a commercial library of 15.5 million molecules (ENAMINE Real Space) identified three novel scaffolds with a potency 100-200 µM. Further derivatives of molecules are continued to improve the potency and physicochemical properties.
After 1 year, Vigneshwaran has achieved the following goals:
- As an initial step, a similarity search using FTrees with three of the potent inhibitors reported in the literature and also characterized in our group was performed. The FTrees approach provided an efficient search from the 15.5 million molecules of ENAMINE Real space and found 80,505 as hit molecules with a threshold of 80% similarity. The resulted hit molecules were further screened using a machine learning approach by applying Random Forest and RDKit Fingerprints. The screening has yielded 2424 molecules from the ligand-based approaches.
- The SeeSAR tool helps to understand the energetics of important structural features of the reference compounds that are required for the inhibition of human CD39. As we apply a homology model of CD39 a detail understanding of the interaction is highly required. Then, in the next step the 2424 molecules identified from ligand-based approaches were screened using the structure-based approach, molecular docking using SeeSAR. By analyzing the estimated binding affinity identified from SeeSAR and the interaction of molecules with the key amino acids in the binding pocket, a total of 516 molecules were selected. In the final step, 30 diverse set of molecules with specific structural features, patterns were selected using MONA and SMARTS Editor. Finally, 21 molecules were purchased based on their commercial availability and affordability.
- The final 21 purchased compounds were investigated using malachite green assay established in our group. The molecules were tested at two concentrations (100 and 200 µM) each in triplicates using ATP as substrate for the human CD39. The screening of molecules found three novel molecules with a potency value of 113, 146 and 226 µM, respectively. The identified hit molecules were compared with other molecules using SeeSAR and helps to understand the binding mode and further designing the derivatives of the molecules with the support of CoLibri.