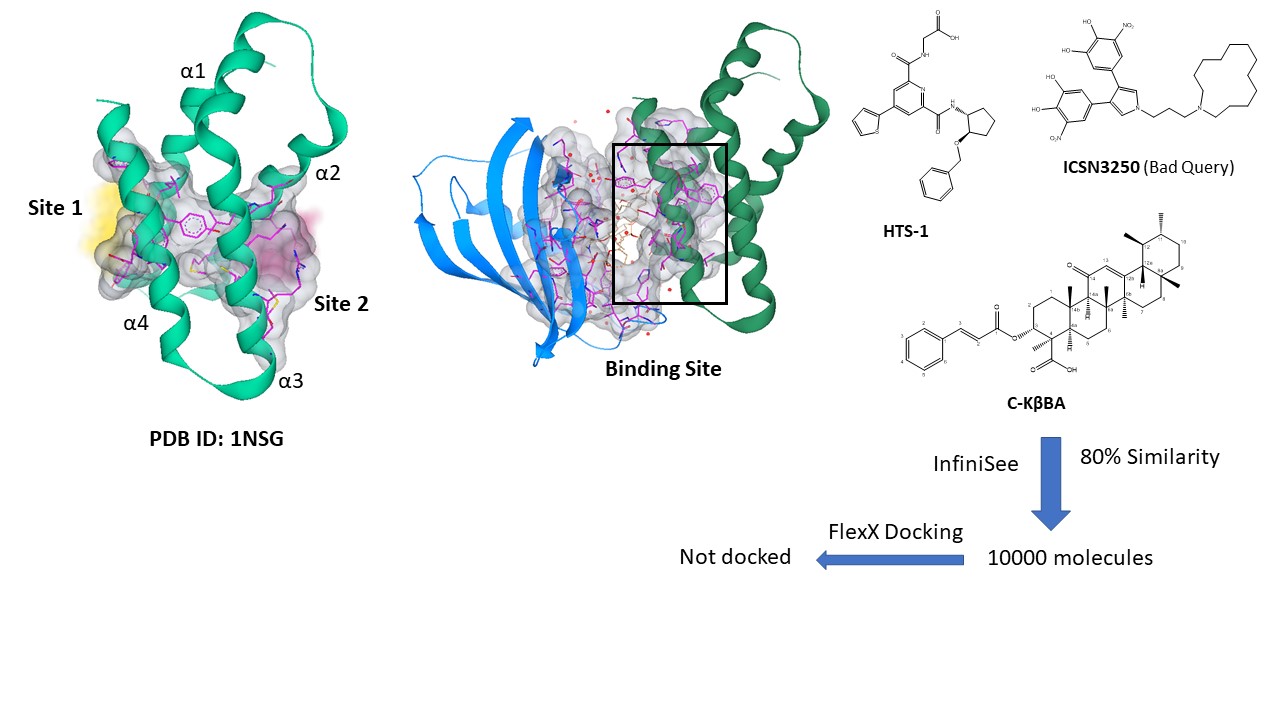
Out of various reported crystal structure of FRB domain of mTOR, X-Ray crystal structure of FRB domain in complex with Rapamycin (2.20 Å resolution, PDB ID: 1NSG) was selected for predicting the possible binding sites. Using binding site analysis mode of SeeSAR, two binding sites were identified in the region between α1-α4 (site 1) as well as α1-α2 helices (site 2). The amino acid residues involved in binding site 1 were similar to those contributing towards phosphatidic acid (PA) binding as characterised by Veverka et al., 2008 and Yoon et al., 2011 using NMR. Detailed investigation of literature provided availability of few experimental inhibitors at early development stage that compete with PA for binding to the FRB domain at site 1. InfiniSee based similarity search of reported inhibitors as reference structure from a library of GalaXi, REAL Space provided 10000 ligands, further docked into site 1 of FRB domain of mTOR.