Pathogenic bacteria resistant to current first-line antibiotic treatments in the United States are responsible for two million infections a year, placing a strain upon the United States healthcare system and increasing expenditure by an estimated $20 billion per year in associated medical costs. Of particular concern is the pathogenic bacterium methicillin-resistant Staphylococcus aureus (MRSA), which exhibits resistance towards almost all clinical β-lactams (e.g. penicillins, cephalosporins, and carbapenems). MRSA commonly acquires additional resistance determinants, giving rise to multidrug-resistant MRSA subtypes (MDRSA) which can be especially challenging to treat. The bacterial
cellular division protein Filamenting temperature-sensitive mutant Z (FtsZ) represents a novel and attractive target for combating such multidrug-resistant infections, with efficacious preclinical allosteric FtsZ inhibitors of the benzamide family reported in literature. However, benzamide-resistant MRSA FtsZ isolates (G193D, G196S, N263K)
hinder application of this compound class, with N263K-mutant FtsZ inducing steric occlusion to the benzamide pharmacophore and conferring resistance to all presently known members of the benzamide family. Accordingly, there is a compelling requisite for the discovery of novel inhibitors of FtsZ which are unhindered by recognized FtsZ
structural mutations.
To this end, the SeeSAR and infiniSee software packages provided by BioSolveIT were instrumental in permitting an evolutionary virtual high throughput screening methodology that resulted in the identification of three promising lead-like candidate FtsZ inhibitors. All proposed inhibitors exhibit favorable predicted ADMET properties suitable for a lead-like candidate and predicted potencies surpass that of the preclinical benzamide TXA707 against the clinically relevant S. aureus MRSA252 strain and the G196S FtsZ mutant, the most common mutant conferring resistance to the benzamide, TXA707. Additionally, all proposed inhibitors demonstrate robust predicted binding modes unaffected by the G196S and N263K mutations with one of the synthesized compounds demonstrating in vitro antibacterial activity against S. aureus. This lends promise to the potential application of these compounds as potential novel inhibitors of FtsZ for the treatment of resistant bacterial infections.
After 1 year, Benjamin has achieved the following goals:
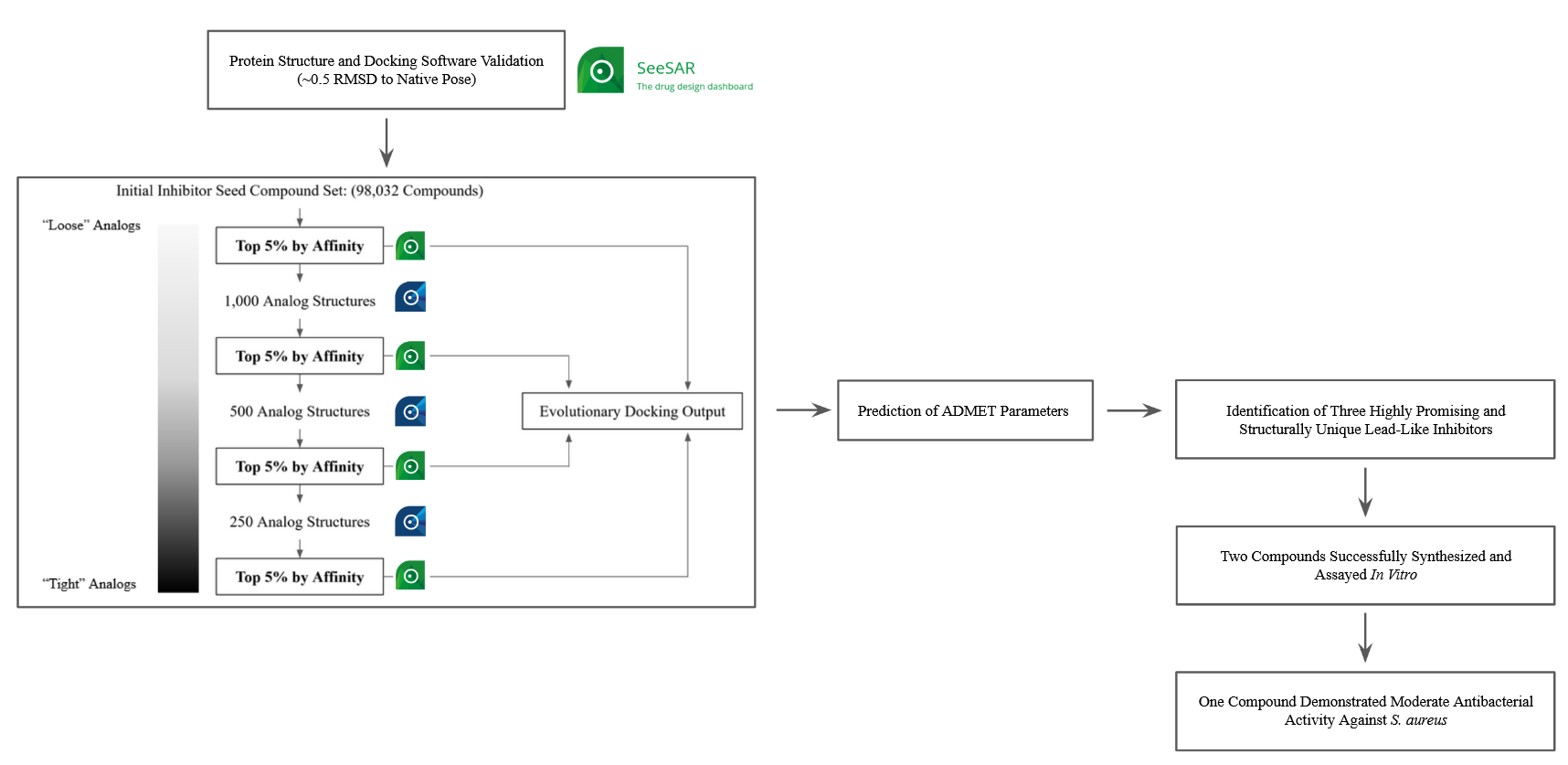
- Preparation and validation of the relevant FtsZ crystal structures (PDB: 5XDT and 5XDV) and docking software [SeeSAR] was undertaken. The relevant FtsZ crystal structures were prepared for molecular docking in UCSF chimera and their associated PDB files were analyzed for structural anomalies in the allosteric binding site of FtsZ with the aid of EMBL-EBI PROCHECK and PDB REMARKs analysis. The validation of SeeSAR for molecular docking was undertaken by introducing TXA707/TXA6101, with no pre-defined bond torsionality, as a ligand to their associated prepared structures, and comparing the predicted binding pose with the native binding pose observed in the crystalline structure through a root-mean-square deviation of atomic positions via a pairwise analysis of all atoms. SeeSAR did exceptionally well in the reproduction of the crystalline binding pose yielding RMSDs of approximately 0.5Å for both structures, validating its predictive accuracy against the target.
- Compounds exhibiting favorable potency and ADME parameters were identified through an evolutionary screening methodology. Known and predicted inhibitors of FtsZ in addition to predicted targets of known FtsZ inhibitors, comprised of 98,032 compounds, formed the initial seed set of compounds. The top 5% of compound by affinity as determined by SeeSAR underwent analog generation in infiniSee to generate analogs that were then subsequently screened by SeeSAR again in a cyclic pattern. This permitted effective scaffold hopping and the exploration of a large section of target-relevant chemical space in a highly time-efficient manner. The resulting hit compounds from this screening had their ADMET properties predicted for the identification of lead-like candidates contained amongst the hit compound set.
- Proposed compounds were synthesized and biologically assayed. Two of the proposed compounds were successfully synthesized, purified, and structurally confirmed via 1H NMR and LC/MS. The first compound was assessed for biological activity via agar disc diffusion with S. aureus and E. coli but exhibited no antibacterial activity against either. The compound was re-assayed as a sodium salt and as a methyl ester, both of which exhibited no antibacterial activity. On account of the lack of antibacterial activity, no FtsZ-specific assay was conducted for this compound. The second compound was evaluated for biological activity via agar disc diffusion with S. aureus and demonstrated a moderate zone of inhibition approximately 10mm in diameter on Mueller Hinton agar plates. However, due to time constraints imposed by COVID-19 no further biological assays on this compound were able to be undertaken in this timeframe.