Since the first time it was observed in the late 2019, COVID-19 spread as very dangerous pandemic which led to a new emergency state all over the world and, moreover, it has become in an issue for all the health care systems. There is a much-needed interest of the scientific community to research for novel antivirals to overcome vaccine drawbacks, virus resistance and, future spreads of family-related pathogens. In this regard, the study of the SARS-CoV-2 Spike protein and its interaction with human the ACE2 trough the Receptor Binding Domain (RBD) is a cornerstone, because it promotes viral infection and it is a very selective drug target. As such, our objectives were focused in the study of this protein-protein interaction to provide a new source for antivirals molecules trough drug-repurposing strategies. Despite a plethora of computational studies aimed to this target during the last months, no candidate has been reported experimentally, confirming the hard and challenging approach. Using the software provided by BioSolveIT, we have iteratively redesigned our strategies to carry out an exhaustive study of the RBD and its potential to bind drug-like molecules to act as antivirals. First, we screened the receptor binding motive by means of virtual screening approaches to find potential candidates. However, our results together with the lack of solid experimental evidences lead us to conclude that this interface is not potentially druggable by small molecules. Then, we turn our attention to the research of small molecules binding pockets on the RBD surface using SeeSAR tools. After applying software in crystal structures (PDB ID: 6M0J) and, in our own structures generated by molecular dynamics (MD) we identified up to 3 potential pockets. Indeed, one of them was then reported as a fatty-acid binding pocket in November 2020. With this result we performed several docking experiments using drug molecule libraries and antiviral compounds (in-house collection from a collaborator). Finally, we selected some candidates to perform MD simulations and further testing in biophysical experiments. As result, we have identified molecules, that are able to bind to the RBD, validating our computational workflow. Moreover, we have discovered that these molecules are also capable to alter the affinity of RBD toward ACE2, confirming its utility to act as antivirals and/or chemical probes Thus, this investigation represent the first-in-class work using computational approaches to identify molecules against RBD-ACE2 successfully, and we will report our breakthroughs to the scientific community to facilitate future investigations on this virus.
After 1 year, Javier has achieved the following goals:
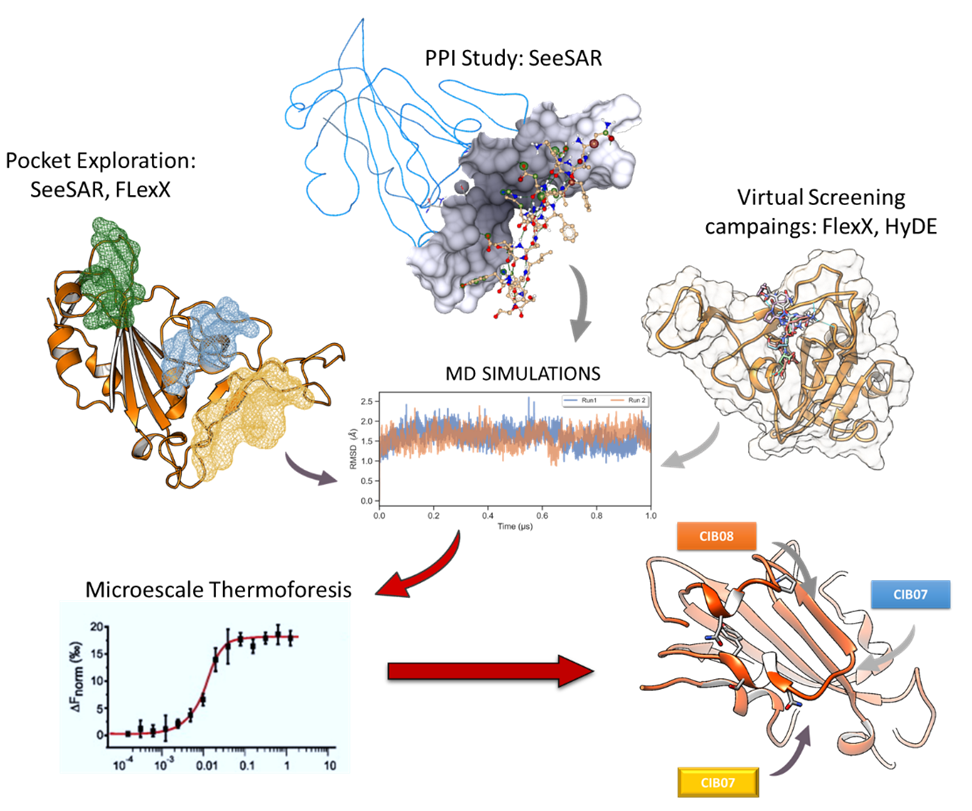
- We have set the basis for prospective docking experiments by deeply studying the RBD-ACE2 interaction interface. Using SeeSAR, the protein editor mode and Hyde, we characterize those residues present in the ACE2 alpha helices which could be representative hotspots for the interaction. These results were combined to other generated by means of MD simulations and MMGBSA methods. Then, we performed several docking experiments with three different libraries of compounds using FlexX. However, poses and scores obtained let us conclude that this might not be a promising druggable site.
- We have explored the presence of cavities and pockets in the RBD which would be able to bind drug-like molecules. Using the binding site mode and its cavity detection functionality, implemented in SeeSAR, in combination with other tools to characterize them we proposed two new binding sites. Subsequently, we carried out MD simulations of the RBD to generate snapshots with different conformational states. Applying these same approaches, we discovered a new site inside the RBD. This site was also described in November 2020 by an experimental work, thus supporting our hypothesis and workflow.
- We have performed virtual screening campaigns in all the detected sites using FlexX and other docking tools. After scoring with Hyde and thorough visual inspection we selected some candidates to perform MD simulations. Those compounds which exhibited a good result also in MD were purchased and tested in microscale thermophoresis against RBD by our collaborators, and in competitive assays. We have identified a few drug candidates able to bind this protein modifying its affinity for ACE2, thus confirming our protocol. Additionally, we observed that the combination of FlexX and Hyde was able to predict more active hits compared to other docking programs used in parallel. Phenotypic assays confirmed that, some molecules exhibited antiviral properties in the assay conditions. Moreover, a second library of small molecules was explored with similar results. Hits thus obtained in this second library will be optimized in a future medicinal chemist program to provide potent antivirals.