S. aureus' DNA Gyrase is a heterotetrameric protein composed of two A subunits called DNA gyrase A (Gyr A), and two B subunits called DNA gyrase B (Gyr B). The main role of Gyr A is cleaving /rejoining of the bacterial DNA molecule during the replication process, while Gyr B hydrolyzes ATP providing the necessary energy for the Gyr A-DNA activity. Currently, there is a much-needed interest in developing novel antibacterial compounds, especially those against strains of multidrug resistant bacteria (MDRB) such as S. aureus. DNA gyrase is a particular important target for inhibition since this enzyme is not present in human beings.
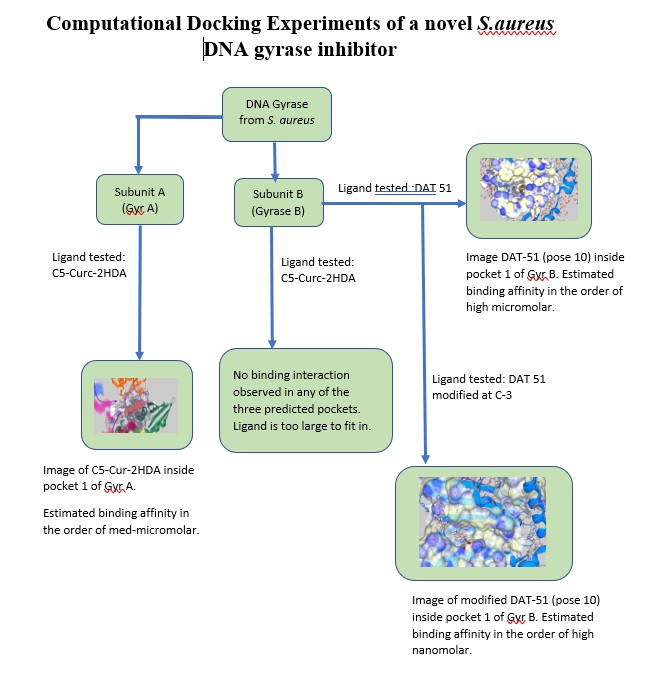
In the first three months of this project we used SeeSAR’s capabilities to explore the possible binding sites of a novel inhibitor (a curcumin derivative called C5-Curc-2HDA) synthesized by Sanabria et al. on S.aureus' DNA Gyrase. From our docking experiments with Gyr A we discovered several possible binding sites. Using SeeSAR' s HYDE estimated affinity calculation, we obtained a low micromolar estimated affinity with the curcumin derivative. In addition, docking experiments showed that this inhibitor is too big to bind to the Gyr B subunit.
In the second phase of this project, we focused on the Gyr B subunit. During this period, we decided to investigate another novel compound from Sanabria's group, in this case an unsaturated fatty acid inhibitor called DAT 51. Using SeeSAR’s docking function, we discovered three possible binding pockets in the Gyr B subunit, the best one (pocket 1) having a total of 23 amino acids. Following SeeSAR’s protocols, pocket 1 was defined as the binding site for docking of DAT-51. A total of 24 poses was generated, affinity calculations for pose 10 showed the best interactions with the residues inside the active site, with a high micromolar to low nanomolar estimated affinity. Using pose 10 conformation as a molecular target and SeeSAR's visualization tools, we identified the presence of unoccupied spaces inside pocket 1. One of these spaces correspond to a hydrophobic space as seen in the binding site surface. To improve affinity, we decided to use the Molecule Editor mode to modify the ligand’s structure, such that this specific empty space was filled with hydrophobic functional groups. After several experiments, the addition of two new methyl groups to carbon 3 of DAT 51 showed the best improvement in affinity of one order of magnitude, from high micromolar range to high nanomolar range.
We were able generate computationally a completely new DAT 51 derivative with an estimated binding affinity in the order of nanomolar to the ATPase activity subunit of S. aureus.
After 1 year, Antonio has achieved the following goals:
- We were interested in docking experiments of novel S.aureus DNA gyrase inhibitors. During this year a new compound from Sanabria's group: an unsaturated fatty acid inhibitor called DAT 51 was investigated. Using SeeSAR, we discovered three possible binding pockets in Gyr B. The best one was investigated using SeeSAR’s software protocols. Defining pocket 1 as the binding site for docking of DAT-51, 24 poses were generated. From estimated affinity calculations and visual inspection of the conformations, pose 10 was chosen. Applying visualization tools to pose 10, we identified the presence of a hydrophobic unoccupied space inside pocket 1. To optimize the estimated affinity of the ligand, we decided to use the Molecule Editor mode to modify the ligand’s structure, such that this specific empty cavity was filled with hydrophobic groups. We found that the addition of two new methyl groups to C-3 of DAT 51 showed the best improvement in affinity from micromolar to high nanomolar range.
- We were interested in locating the binding site of our S. aureus’ DNA gyrase inhibitor, C5-Curc-2HDA . We were able to identify possible binding sites in Gyr A and Gyr B. In the first phase of this project we used SeeSAR’s software to dock our curcumin derivative in the binding sites identified in phase 1. To our surprise none of the binding pockets identified in GyrB could bind the curcumin derivative, visual inspection indicated the inhibitor was too big to occupy any of the binding pockets. For Gyr A we were able to demonstrate that the curcumin derivative can bind to the pockets predicted by SeeSAR, which could explain the antibacterial activity observed experimentally.
- Following our results from phase 1, we decided to explore further the binding pockets in Gyr B with smaller compounds. Since the curcumin derivative was too big for the binding sites of Gyr B, we used another inhibitor of DNA Gyrase: a fatty acid derivative called DAT 51 (372 amu) from Sanabria’s group. We demonstrated that this inhibitor binds to one of the three pockets identified by SeeSAR with a low micromolar estimated affinity.