The mitochondrial protein dynamin-related protein (Drp1) has been implicated in the development of a number of neurodegenerative diseases, including Alzheimer’s disease. To date, no direct small molecule inhibitors of human Drp1 have been identified. A year ago, we started this project with three "hit" compounds (OB-27, OB-37 and OB-47), from three alternate chemical classes, which interacted with Drp1 directly and inhibited its activity in cell-based assays. Using FTrees to search for compound analogues, we found 100 analogues to purchase/synthesise for each compound class. Using SeeSAR to dock and rank the compounds, we purchased 64 analogues.
Surface plasmon resonance (SPR) assays showed that 19 of these compounds directly interacted with Drp1 in a dose-response manner (Kd range 3-520uM, notably 5 compound were insoluble and 13 compounds bound to SPR chip thus binding affinity could not be assessed). This gave us an adjusted hit rate (taking into consideration compounds which could not be assessed) of approx 53%. Importantly, 12 of these interacting compounds were from the same chemical class (OB-47 analogues). This result allowed us to focus our chemistry effort into this class of compounds.
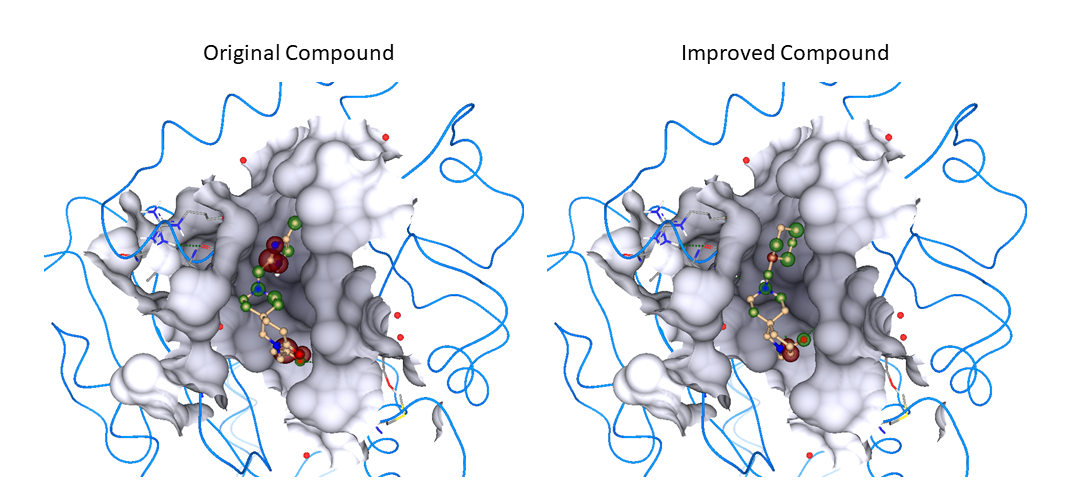
Using SeeSAR, we then designed 20 compounds predicted to have better activity than our lead compound, to synthesise and assay. Unfortunately, due to COVID-19, these studies have not yet been completed.
In summary, BioSolveIt software assisted us in choosing compound analogues to purchase (giving an adjusted hit rate of approx 53%). The biological evaluation for these analogues led to a front runner compound series. BioSolveIT software was again used to design compounds of this series with improved activity.
After 1 year, Jessica has achieved the following goals:
- Search for chemically similar compound analogues to purchase. FTrees was run, in the infinisee interface using the REALspace_2019-05 library on our top 3 compounds classes. The top 100 analogues were shown for each compound. I removed any compound analogues that I have already purchased and tested.
- Dock all compounds into our crystal structure and rank the best before purchasing Compounds were docked and analysed in SeeSAR, resulting in the purchase of 64 analogues. These were assayed giving an adjusted hit rate of approx 53%.
- Identify and improve a front runner compound series for medical chemistry effort. Assays identified a clear front runner series (13 active compounds). SeeSAR was then used to design 20 analogues for chemical synthesis.