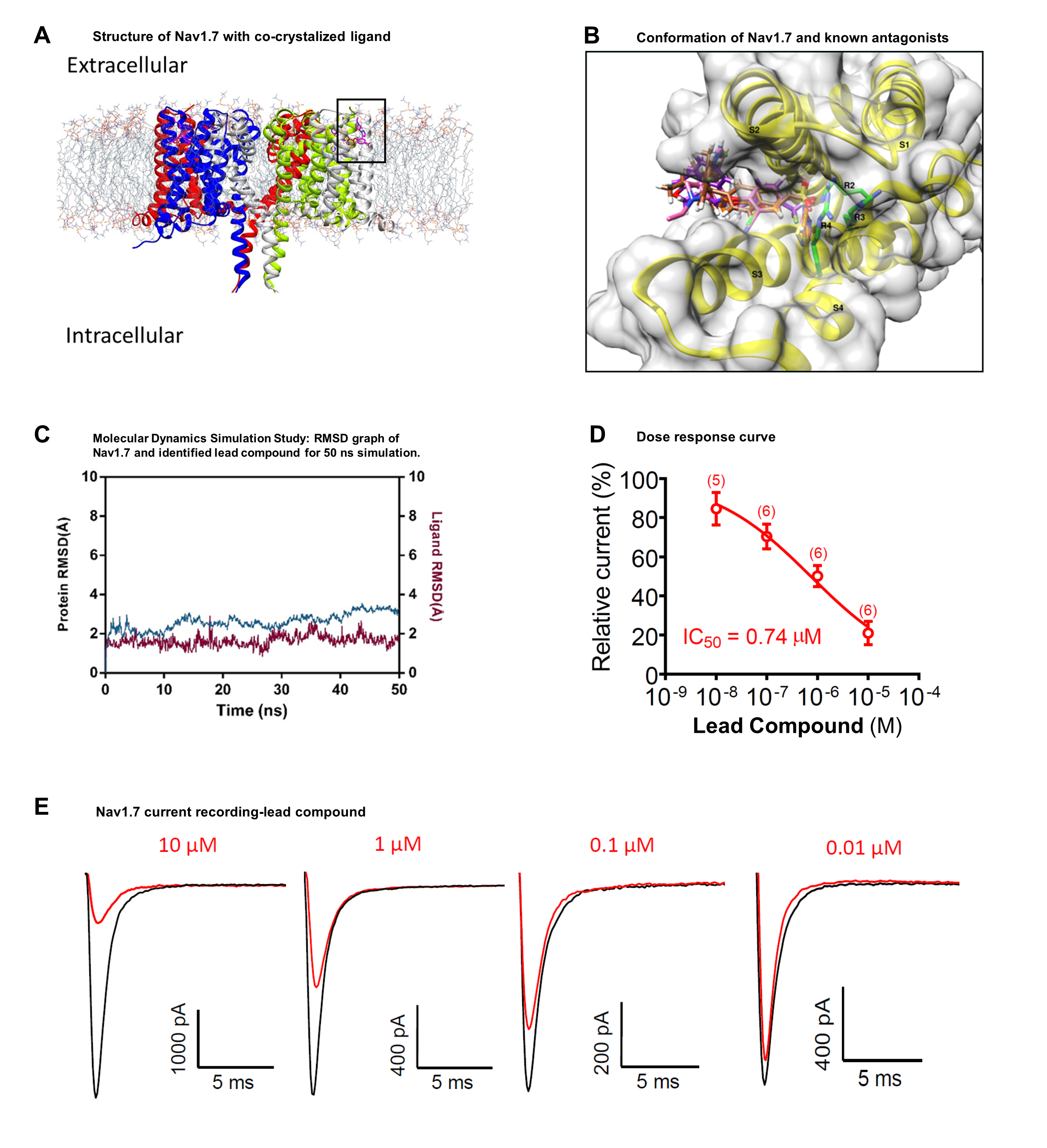
Since last 12 months, with the help of Biosolveit software we have identified a novel chemical scaffold which acts as a potent inhibitor of human voltage-gated Na+ ion channel Nav1.7 for the treatment of pain. Chronic pain (CP) is a disease that results from an injury or a disease that affects the somatosensory nervous system. CP is gradually becoming a serious global health problem. Approximately 10-55% of the world’s population is suffering from chronic pain. In addition, the cost of treating chronic pain is enormous, which imposes a huge burden on health budgets. Therapeutic approaches for chronic pain have limited effectiveness. As a result, physicians prescribe opioids for chronic pain, which leads to an epidemic of opioid prescription, abuse and addiction. Thus, the development of novel, effective and safe drugs for CP is still an unmet medical need. Voltage-gated sodium (Nav) channels act as molecular targets of cardiovascular and neurological disorders. It is difficult to design selective inhibitor of Nav channels because there are nine closely related isoforms (Nav1.1 - 1.9) that share high sequence identity. We are targeting the Nav1.7, which is found in the peripheral nervous system and involved in pain perception. A new class of analgesics targeting voltage-gated sodium channels (Nav) could help to treat people with CP. Before applying for the Biosolveit challenge we obtained some preliminary data, in which we have standardized molecular dynamics simulation (MDS) experiment with positive and negative control and a few docked compounds from a small compound library. Then our next objective was to screen 1.5 million compound library for identification of inhibitor for Nav1.7 with analgesics effect. We have developed this project with the computational support of Biosolveit software. In this study, we designed a protocol for the identification of isoform-selective inhibitor of Nav1.7, by utilizing the class of isoform-selective antagonist data. Initially, a similarity search was performed and the identified hits were docked into a binding site on the fourth voltage-sensor domain, VSD4. We used the FTrees tool for similarity searching and library generation, the generated library was docked in the VSD4 domain binding site using FlexX and compounds were shortlisted using a FlexX score and SeeSAR hyde scoring. Finally, the top 25 compounds were tested with MDS. On the basis of MDS RMSD plot we have narrowed down our list to 9 compounds and purchased them for in vitro and in vivo validation. These experiments have led to identify a novel compound with good activity and a very interesting IC50 value (0.74 µM). With same cycle of above methods, we have started optimization of our novel lead compounds.
After 1 year, Sharat has achieved the following goals:
- ‘Accomplished Virtual Screening and in silico validation’. The database containing 1.5 million compounds was utilized for in silico similarity searching. We used known Nav antagonists PF-05089771, and GX-936 as query molecules against compound database. Therefore, a library of 2000 compounds was designed based on the Global similarity score as well as local similarity score of aryl sulfonamide group. It was shown in previous studies that the isoform specific inhibitor can be designed by targeting the non-conserved residues found on the S2 (YWxxV) and S3 (GMxxA) transmembrane helices. Therefore, the active site for docking experiments was delineated by selecting these residues and the generated library of 2000 compounds were docked into this site using FlexX module. The compounds were also assessed for ligand affinity with HYDE in SeeSAR for further validation. After analysis, a list of 25 compounds was prepared to perform MDS.
- ‘Hit identification’. The binding site for Nav1.7 is partially surrounded by membrane bilayer. It has been observed that a bound phospholipid forms a trimeric complex with the inhibitor and the channel protein. Therefore, we have performed a series of membrane simulations of Nav1.7 in complex with identified compounds. These complexes were subjected to 50 ns simulation using OPLS-2005 force field. The final Nav1.7-compound complex of each MDS setup were re-evaluated by SeeSAR and HYDE. The complexes whose values for ligand RMSD were lower than the protein RMSD were selected for further assessment. Based upon these analysis, we have purchased a set of 9 compounds to undergo biological evaluation.
- ‘In vitro and in vivo confirmation of compounds’. Whole-cell patch-clamp recordings in HEK-293 cells and DRG neurons were conducted. We used patch pipettes to record transient Na+ currents. One of the compound dose-dependently reduce the peak sodium currents in Nav1.7-HEK-293 stable cell line, with an IC50 values at 0.74 µM. At 10 µM concentration of lead compound has a weak inhibitory effect on the sodium currents in both mouse and human DRG neurons. We also tested whether lead compound could attenuate formalin-induced inflammatory pain following spinal intrathecal route via lumbar puncture. Intrathecal injection of 10 nmol has a weak anti-nociceptive effect in the second phase of formalin test. Our findings suggest that lead compound have some effect in reducing pain. We plan to proceed with optimization of the lead compound to improve its efficacy and to reduce the IC50 values.