During last 12 months, we have been working in a new project about protein-protein interactions (PPIs). We have developed the project with the computational support of Biosolveit software. Chronic kidney disease (CKD) is the non–transmissible global cause of death that raised the most within the past 20 years. It increases the risk of all–cause mortality and may progress to end–stage of renal failure. There is no effective therapy for CKD and current approaches do not prevent its progression. Then, the search of new and validated targets against CKD has become a milestone in academic groups and industry. CKD has been related, at least partially, to integrin–linked kinase (ILK). This is an intracellular pseudokinase which forms the ternary complex IPP (ILK–PINCH–parvin) with two adaptor proteins. On the other hand, disrupting PPIs has emerged as a very promising strategy against different types of diseases due to its high selectivity and specifity. We have focused our efforts in disrupting the ILK–parvin interaction to validate this protein as target for CKD and to get insight of the function of this complex. Our first effort was aimed to identify the hotspot of the surface of interaction in ILK. We were able to identify a hotspot, with the help of Seesar and HYDE, in a new way which agrees with other representative methods. Once we acquired this information, we prepared a set of small peptides and optimized them using ReCore in order to validate this proof of concept. As a result of the former, some of these peptides exhibited the desired activity in phenotypic assays, thus supporting our hypothesis. Next, we prepared longer peptides to get more insight onto the PPI. We used Seesar to study all the peptides resulting from an alanine scanning, and Flexx to dock all of them. Again, we used HYDE scoring function to correlate HYDE energy values with affinity values (KD) obtained by SPR experiments. These results have let to identify a peptide with good activity and a very interesting Koff value (0.691 uM), which could be used as tool in biological chemistry. Finally, we are now performing a virtual screening using ZINC database and a consensus approach to identify peptidomimetic structures. Thus, we are using HYDE as a scoring function for the best docked candidates in order to select them for further steps of virtual screening (i.e. molecular dynamics) and synthesis. Summarizing, we have proved the versatility of Biosolveit software to lead new projects in the field of PPIs never studied before. Moreover, our results will be completed by identification of small hit molecules as potential drug-like compounds able to disrupt ILK-parvin interaction.
After 1 year, Javier has achieved the following goals:
- Identification of Protein-Hotspot. Starting from the crystal structure of ILK-parvin and the sequences of both proteins, an extensive analysis on the interaction surface was carried out using different in silico approaches to identify the most favourable regions of interaction. Particularly, we used Seesar and HYDE to characterize the interacting peptide of parvin onto the ILK surface. As a result, we were able to identify a hot-spot in ILK which is in a very good agreement with others approaches such us FTMap, Hot-Spot Wizard or Fragment Hot-Spot Maps. This hotspot corresponds to a hydrophobic interaction established by the Phe307 residue located in parvin which is well buried into ILK surface. This knowledge, as well as the energetic per-residue contribution was the basis of our further designs.
- Design and characterization of small peptides able to inhibit ILK-pathway in vivo. Based on our previous milestone, we designed a small set of tripeptides with the help of Seesar and ReCore. Seesar allowed us to explore regions of the interaction interface and to prepare novel peptide using rational design. Our Hit compound was the tripeptide Ac-Tyr-Phe-aa3-OCH3. Once the design step was accomplished, all the peptides were evaluated by docking with Flexx and HYDE in order to select the most promising ones for further synthesis. As a result, we have found some hits which exhibit a good in vivo profile by inhibition of AKT and GSK phosphorylation (two enzymes involving in ILK signalling), and cytoskeleton destabilisation.
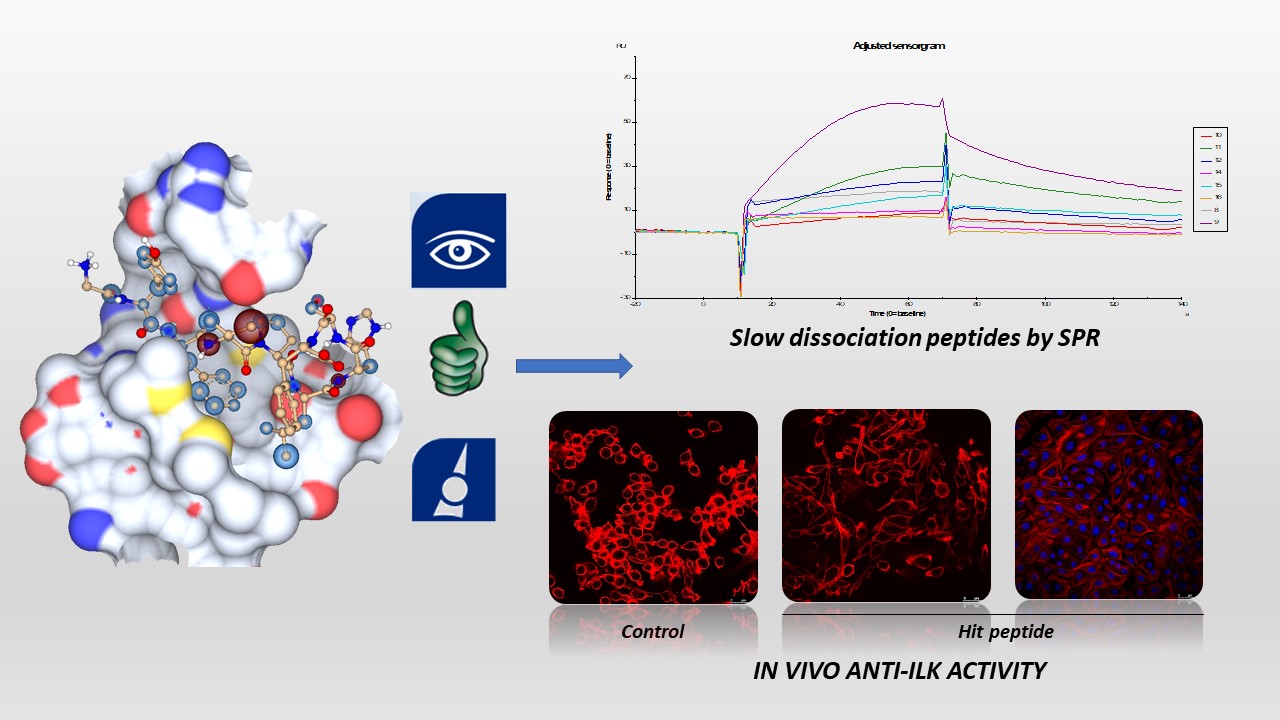
- Study of interaction surface by in silico and in vitro approaches. Based in our results regarding with hotspot identification and small peptide activity, we proceeded to study the full interaction surface. For this aim, we used Seesar to get insight into the per-residue energetic contribution of parvin peptide epitope to the ILK interaction surface. Assisted by the straightforward visualization of the results of SeeSar, we performed an alanine scanning both in silico and in vitro, and we designed peptides with different lengths which were docked and scored with HYDE. We prepared these longer peptides and we studied them by Surface Plasmon Resonance (SPR). As a conclusion, we have identified a set of promising peptides with slow dissociation kinetics (low Koff) which will be used as synthetic tools to study ILK-parvin interaction. Currently we are trying to prepare some new fluorescent probes using these peptides.