This study aimed to identify novel hit compounds targeting soluble epoxide hydrolase (sEH) for the development of lead molecules that could be used in the treatment of chronic inflammatory diseases. While numerous sEH inhibitors have been reported, none have reached clinical use due to high plasma protein binding and suboptimal pharmacokinetic profiles. The sEH enzyme regulates inflammation by converting protective endogenous epoxylipids produced through cytochrome-mediated epoxidation of fatty acids into inactive diols. Thus, there is a significant therapeutic potential for small compounds that selectively inhibit sEH to retain these epoxylipids.
We targeted the identification of small molecules exhibiting strong sEH inhibition with favourable ADMET properties by integrating artificial intelligence–driven and physics-based strategies. To this end, millions of stock molecules, clinically approved drugs for repurposing, fragment libraries, and billion- to trillion-scale chemical spaces (GalaXi, CHEMriya, AMBrosia, Freedom Space, REAL Space, and eXplore) were screened using structure-based, pharmacophore-guided Chemical Space Docking (CSD). The method enabled efficient coverage of extensive chemical spaces, completing some smaller libraries in hours and the larger ones in a matter of weeks, surpassing what conventional structure-based virtual screening methods could achieve. For each
screening workflow, the most appropriate screening
parameters were defined through multistep systematic analysis. Then each model's performance is evaluated through active and decoy datasets and optimized by fine-tuning model parameters. With validated protein structures and screening models, large-scale virtual screening was carried out on high-performance computing (HPC) systems, including our internal cluster Chimera, the Turkish Science e-Infrastructure (TRUBA), and MareNostrum V at the Barcelona Supercomputing Center.
Early hit compounds were analysed using molecular dynamics (MD) and MM-GBSA to prioritize compounds based on key residue contacts and predicted binding affinity. A total of 291 compounds were selected for experimental testing. The assays at the very early stages has already revealed activities ranging from <0,01 µM and 3 µM. The studies are still ongoing. Additionally, subsequent enzyme kinetics and ITC studies will assess the inhibition mechanism binding thermodynamics and X-Ray crystallography or CryoEM. Thus far, the findings show that our integrated computational and experimental pipeline can identify potent, experimentally validated sEH inhibitors with drug-like properties, providing candidates for translational and early preclinical development.
After 1 year, Emine Yekta has achieved the following goals:
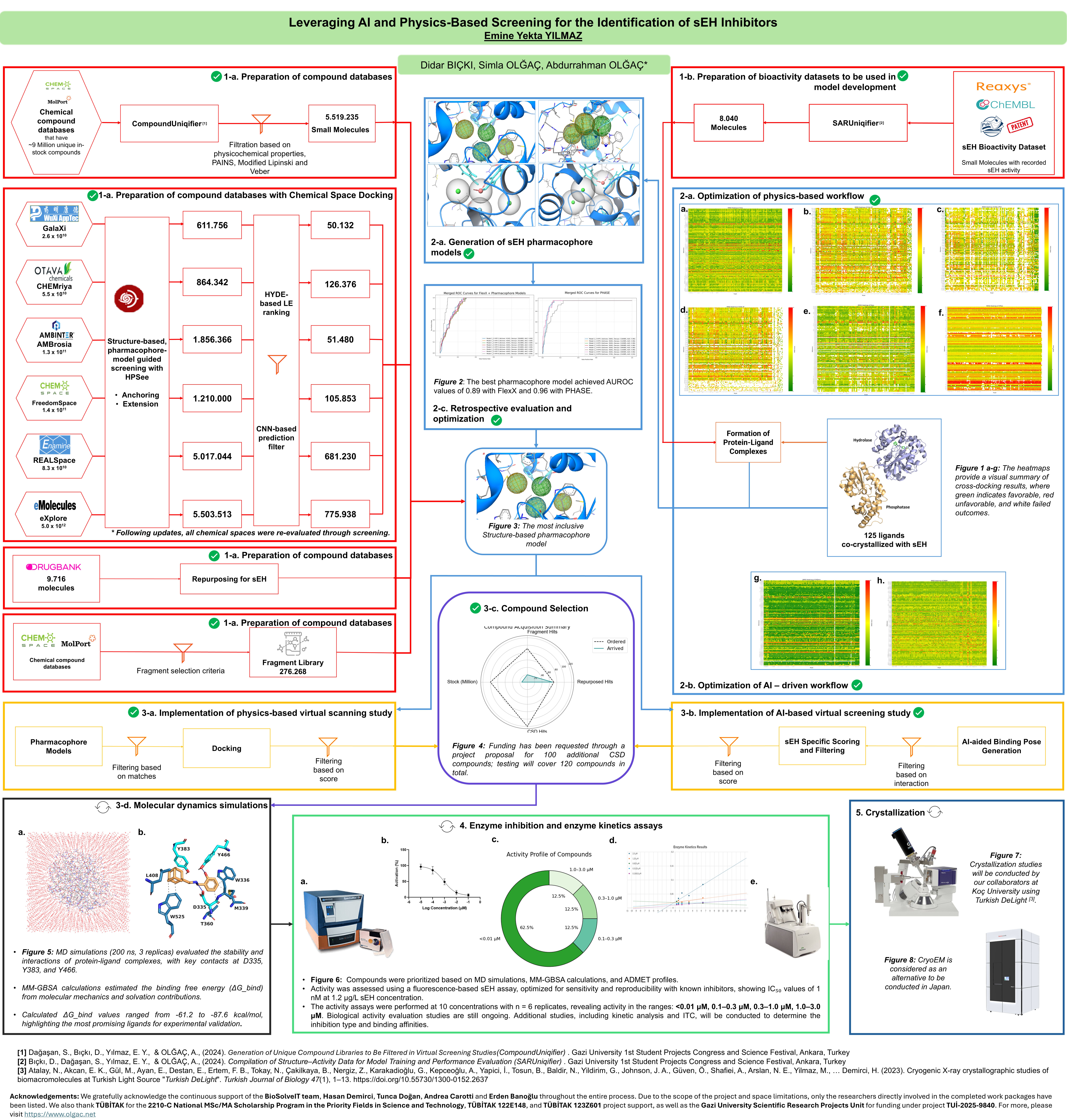
- This stage aimed to prepare well-curated compound databases and implement AI and physics-based screening workflows for sEH inhibitor discovery. A bioactivity dataset was curated with 8040 high-confidence data points, which was later used to identify actives and generate decoys to support systematic cross-docking and the generation and validation of pharmacophore models. A chemically diverse and computationally manageable small molecule library was prepared, and filtered, reducing the in-stock compound library to 5519235 candidates. In addition, 9716 DrugBank compounds and 276268 fragments were screened. Physics and AI-driven screening pipelines were then optimized and executed. Chemical Space Docking and CNN-based screening methodologies were applied across six diverse billion to trillion scale chemical spaces, which yielded 1791009 purchasable hits. The compounds were started to be purchased which were expected to yield with strong predicted binding and drug-like properties.
- Next, promising compounds were selected and their binding mode characterized using MD. CSD outputs were ranked by score and then subjected to cutoff and CNN-based filtering. Remaining compounds were clustered to identify representative hits, and ADMET profiles were estimated to ensure drug-like properties. Each selected molecule underwent 200-ns MD simulations in triplicate following a standard relaxation protocol. MM-GBSA calculations were performed, with ΔG_bind values ranging from –61.2 to –87.6 kcal/mol, supporting prioritization of ligands for biological evaluation. The resulting binding modes with sEH were analysed in detail, focusing on interactions with key residues D335, Y383, and Y466. A total of 291 compounds were advanced to experimental testing, including 100 from stock libraries, 78 from DrugBank repurposing, 91 from fragments, and 20 from CSD results. Funding sought to evaluate 100 additional compounds obtained from the CSD study.
- The following step in our pipeline involved in vitro validation of selected compounds and evaluation of their biochemical activity. A fluorescence-based sEH assay was optimized and validated for sensitivity and reproducibility using known inhibitors. Purchased compounds, prioritized based on MD simulations, MM-GBSA calculations, and ADMET profiles, were screened across ten concentrations, followed by IC₅₀ determination. Wet-lab experiments are ongoing, with completed assays showing activities ranging from <0,01 µM to 3 µM. This step bridges computational predictions with experimental measurements, confirming the relevance of the selected hits. Subsequent studies will include enzyme kinetics to elucidate the inhibition mechanism, ITC to quantify binding affinities and thermodynamics, and structural biology studies to resolve the inhibitor–protein interactions at atomic resolution.