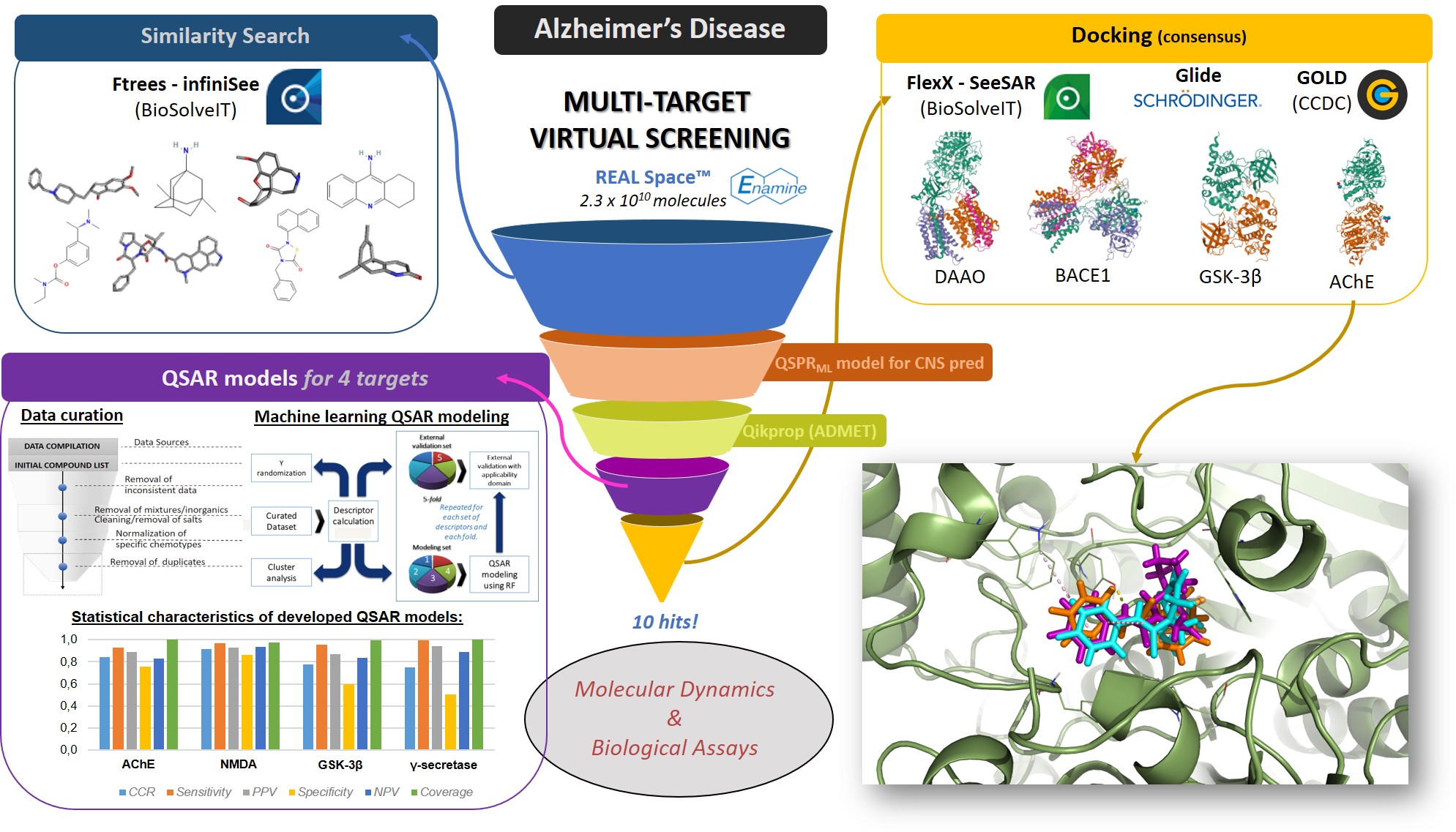
To begin with, I’d like to express my gratitude to BioSolveIT. Participating of this 12-month scientific challenge has been remarkable and allowed me to conduct my own proposal with independence and responsibility. Importantly, this work was only possible in view of the great speed, quality and elegance of BioSolveIT’s software. My proposal was related to my PhD theme, however directed to performing a multi-target (MT) virtual screening (VS) for Alzheimer’s disease (AD). It is clear that AD represents a severe problem to current society that has not an effective treatment available yet. Also, AD shows a multi factorial nature and there are several validated targets associated with its pathological patterns. MTVS has thus become a promising trend in CADD. Despite its attractiveness, so far this approach hasn’t resulted in the approval of effective drugs. Therefore, we developed a MTVS study including different methodologies and consensus approaches to discover novel AD drug. Initial screening was caried out using FTrees considering 8 known AD drugs as queries to build a small library out from the billionaire REAL chemical space. From this library, we only kept molecules with favorable ADME properties and CNS-like profiles, as predicted by QikProp and our in-house machine learning (ML) QSPR model. Reminiscent molecules were predicted by robust ML QSAR models with regard to their potential to act, simultaneously, as inhibitors of 4 AD-targets. To build each model we compiled large publicly available datasets of inhibitors of the corresponding targets: AChE, NMDA, GSK-3β and γ-secretase. QSAR models were developed using Random Forest and showed the respective CCR: 84, 92, 78 and 75%. Application of such QSAR models in MTVS furnished us 39 compounds with enriched structural diversity. In sequence, these compounds were submitted to docking studies, considering a consensus analysis of scores obtained by 3 different software: FlexX, Glide, and GOLD. We used the targets of AChE, D-amino acid oxidase, GSK-3β, and β-secretase. All targets were extracted from the PDB, previously prepared, and validated by redocking in each software. Compounds were prioritized considering their median rank of scores obtained by each software and inspection of their pose’s superposition. We thus selected 10 compounds as promising hits able to act as multi-target anti-AD agents. Recently, we found out that MT drugs are more likely to be successful when designed to a small number (≥2) of related-targets. Therefore, we are now applying ROBOKOP to elucidate drug–target–disease correlations and prioritize 2 related-targets and respective hits. These will be submitted to MD studies and biological evaluation. Naturally we intend to publish this work elsewhere whenever possible.
After 1 year, Guilherme has achieved the following goals:
- Chemical space exploration and compilation of AD-like library. An intense review of the literature has been done. Publicly available content spanning from articles to specific data related to AD and its targets has been gathered and inspected. This allowed us to, initially, select 8 representative AD drugs: donepezil, dihydroergocristine, galantamine, huperzine, memantine, rivastigmine, tacrine, and tideglusib. These were used as queries to perform similarity search in FTrees-infiniSee, using REAL Space (>23 billion molecules) and the following settings: target similarity 0.90, minimum similarity 0.60, and total diversity 0.90. For each drug, 100 compounds were retrieved, totaling a library of 800 AD-like compounds. We emphasize that such library has a great value, also for future applications, as it represents a reduced chemical space of molecules with similar physicochemical properties but different scaffolds from known anti-AD agents.
- Development and application of ML QSAR models. We build 4 ML QSAR models for the targets: AChE, NMDA, GSK-3β and γ-secretase. For each one of them, we compiled the largest publicly available datasets of inhibitors of the corresponding targets from ChEMBL. Data curation from such datasets was conducted by cleaning biological assays, structure validation and normalization, and deletion of duplicates. QSAR models were developed in KNIME using Random Forest and Morgan fingerprints. Moreover, they were validated following the best practices proposed by OECD, employing a 5-fold external cross-validation, performing Y-randomization, and considering respective applicability domains. The respective 4 QSAR models have shown correct classification rates (CCR) of 84, 92, 78 and 75%. These were applied in the MTVS campaign to filter out only those molecules with potential to act, simultaneously, as inhibitors of 4 AD-targets.
- Docking studies to elucidate interactions and prioritize hits as potential MT anti-AD agents. 3 methodologies/software has been used to conduct docking studies: FlexX-SeeSAR, Glide (Schrodinger), and GOLD (CCDC). We tried to keep the same targets used in QSAR studies, however for both NMDA and γ-secretase, no PDB complexes were available. Thus, we used their corelated enzymes D-amino acid oxidase and β-secretase, respectively; in addition to AChE and GSK-3β. All targets were previously validated by redocking, in each software. To prioritize hits in the MTVS campaign, we considered the median rank of scores obtained by each docking software. Furthermore, we inspected their corresponding congruence of poses and interactions, e.g., Hit 1 showed a great overlap of poses obtained by 3 software at AChE binding site and kept important interactions, similar to the respective native ligand. Hence, we selected 10 compounds as promising hits able to act as multi-target anti-AD agents.