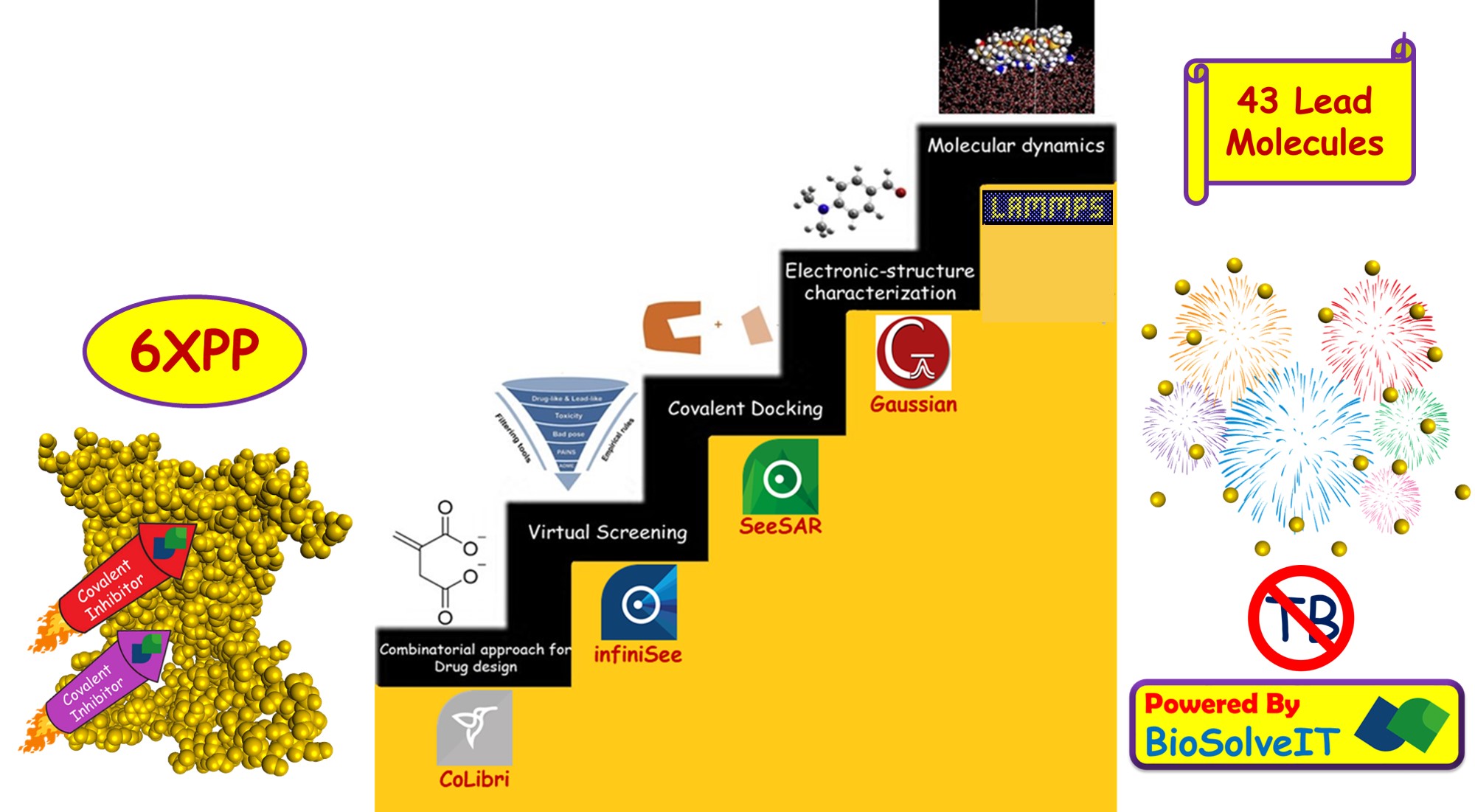
The twelve months of the Fall 2021 scientific challenge went lightning fast. But the efficiency and lightning speed of SeeSAR and infiniSee softwares made the year productive. An internship organized by the Indian Institute of Technology Indore (IIT Indore) caught my attention towards the seriousness of Tuberculosis. During that period, I was working on covalent inhibitors, and motivated by their inhibition efficiency towards various fatal diseases, I started research on utilising the potential of covalent inhibitors towards Tuberculosis. Soon, the Fall 2021 Scientific Challenge of BioSolveIT came on my way to give life to my project. The efficiency of covalent inhibitor, Itaconate against mycobacterium tuberculosis influenced to prepare a series of covalent inhibitors. The covalent libraries provided by BioSolveIT were used for virtual screening. From a total of about 436,457 molecules, seven hundred and seventy nine molecules were selected for the study. Then the selected molecules were analyzed using the analyser feature of SeeSAR to determine optimum properties. Further, these molecules were covalently docked with isocitrate lyase using SeeSAR. The affinity of selected molecules towards the defined binding site was noted and that lead to the discovery of forty three molecules which have lesser inhibition constant when compared to Itaconate. The result excited me and on further analysis, it was found that one of the selected molecules has 1/800 times the inhibition constant of Itaconate. Among the discovered forty three lead molecules, the above mentioned lead molecule has the greatest ligand efficiency as well. Further electronic-structure characterization was done using Gaussian software to gain deeper insights on the reactivity and selectivity of the molecules. For a better understanding of this biological system, a dynamic model suits better than relatively rigid structures came across in the previous analyses. Therefore, molecular dynamics simulation was performed for the lead molecule. MD simulations helped in gaining a deeper understanding of the covalent bond formation and the inhibition process. The major aim of the study was to identify potential inhibitors against mycobacterium tuberculosis. The discovery of forty three lead molecules against the disease, is a catalyst to perform further studies on these molecules. Synthesis and in vitro evaluation of the best molecules among these forty three, can result in successful outcomes in future.
After 1 year, Aaron Kuriakose has achieved the following goals:
- Itaconate, a mammalian antimicrobial metabolite has shown excellent efficiency against isocitrate lyase that causes Tuberculosis. Recognizing the efficacy of covalent inhibitors, the present study was initiated by the retrieval of suitable molecules. Virtual screening was done to obtain covalent inhibitors that are derivatives of Itaconate. This was performed by employing the infiniSee software. The screening was done by making use of the covalent libraries namely Asinex, ChemDiv, Chemspace, Enamine, LifeChemicals and Reaxense. Around seven hundred and seventy nine molecules that have similarity with Itaconate (0.80 to 1.00) were retrieved from 436,457 molecules available in the covalent libraries. Further these molecules were analyzed by utilizing the analyser feature of SeeSAR to determine optimum properties such as solubility, lipophilicity, topological polar surface area and blood-brain barrier permeation. Then these molecules were covalently docked using SeeSAR.
- In SeeSAR software, protein structure of isocitrate lyase was downloaded from Protein Data Bank. Then its binding site was defined for estimating the affinity of screened molecules against the protein. Different poses of the chosen molecules were generated and covalently docked. Further the docked complexes were analyzed for estimated affinity to determine suitable lead molecules. More than forty molecules exhibited lesser inhibition constant than Itaconate. The molecule with most affinity has 1/800 times the KI of Itaconate. In addition, this lead molecule with thiol warhead has the greatest ligand efficiency. Good torsional quality and lack of molecular clashes makes this the prime one. Top ten lead molecules with the best estimated affinity were selected for electronic-structure characterization. FMO and global descriptors analyses were done to enumerate the reactivity and selectivity of molecules. The selected leads showed appreciable reactivity and selectivity towards the target.
- The lead molecule that showed the greatest efficiency has been selected for molecular dynamics simulation using LAMMPS simulator package. The pairwise interactions have been modelled using CHARMM force field. The system has been equilibrated for 5 pico-seconds and production run has been carried out for 300 pico-seconds. The equations of motion have been solved every 2 femto-seconds. The trajectories are analyzed every 30 pico-seconds to identify the changes undergoing in the active site. Molecular dynamics simulation of this lead molecule aided in gaining greater insights about the covalent bond formation and the inhibition process.