At the end of year 2019, COVID-19 emerged and expanded in a flash across the globe setting humanity at the verge of desolation. Healthcare systems were experiencing emergency situation and scientific community was striving to discover new potential antivirals that could encounter the mutant strains. Considering the need, we designed our project in which we have targeted three potential drug targets of SARS CoV-2 namely TMPRSS2, cathepsin and furin. After one year of our careful work, we have successfully discovered potent inhibitors against our targeted proteins to stop the viral entry, replication and pathogenesis at the same time. Precisely, TMPRSS2 and cathepsin inhibitors obstruct the entry of the virus by hindering the cleavage of spike (S) proteins whereas furin inhibitors are responsible for preventing the infectivity and pathogenesis by disrupting the precursor proprotein cleavage and obviating cytokine storm. We have used BioSolveIT softwares and tools to analyze the protein for finding binding sites, searching similarity of inhibitors, modeling pharmacophore, generating and selecting best hits and scrutinizing the optribrium and pharmacokinetic properties of each inhibitors. Throughout our work, core-replacement and fragment growing options of Inspirator mode in SeeSAR have proved to be exceptionally handy and user friendly to discover the lead compounds.
After 1 year, Sumera has achieved the following goals:
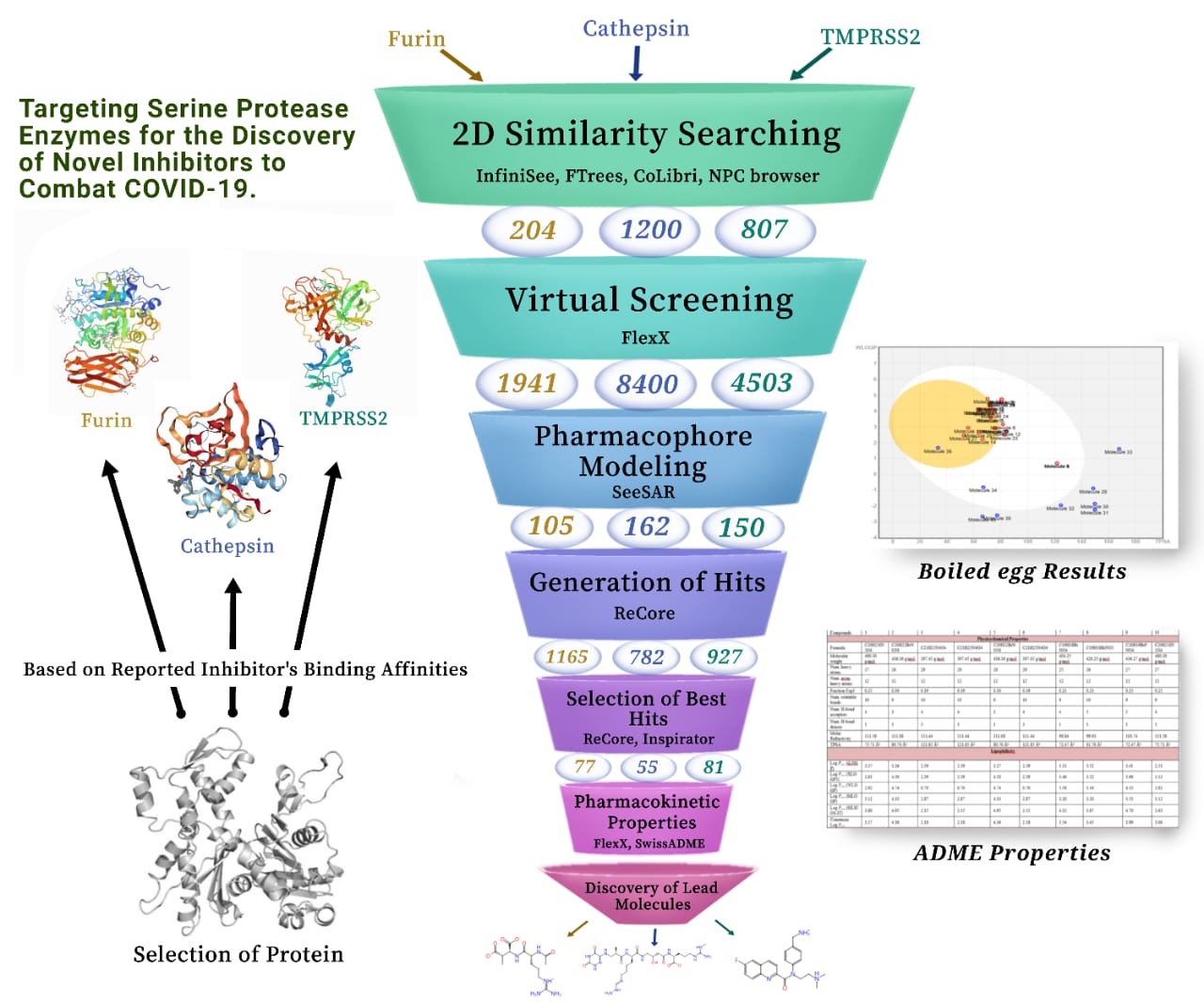
- We have searched reported inhibitors against our targeted proteins from literature and found the similar compounds using BioSolveIT softwares. As we have three target proteins, so we have selected two already reported inhibitors for each. Initially, evaluation of the binding affinities of these reported inhibitors with the binding domains was employed to select the right protein PDB structure. After the selection of proteins, all the binding sites were analyzed and the residues taking part in inhibition were determined using SeeSAR. Subsequently, FTrees was used for 2D similarity searching of each of the inhibitors and generated a number of similar compounds for further analysis.
- The structurally similar compounds were than exported to SeeSAR where they were docked, against their respective protein, generating various poses. Each of the pose was thoroughly investigated for selection via optribrium properties and estimated affinities. After virtual screening of the poses, the remaining molecules were used for the formation of pharmacophore by considering co-crystallized ligand as a reference. The molecules that perfectly align with the pharmacophore were selected and exported to the inspirator mode. In inspirator mode, Recore was used to improve the affinities, remove torsions and clashes from the selected molecules to generate hits.
- The best hits were selected for pharmacokinetic studies via SwissADME and SeeSAR. Some of the molecules were representing remarkable results while others were against expectations. Therefore, the former ones were transferred to molecular editor mode where specific bonds and atoms were added to get the expected results. It was followed by the ADME analysis and the results were evaluated in the form of boiled egg. The best compounds were selected and synthesized to further evaluate their activities in vitro.