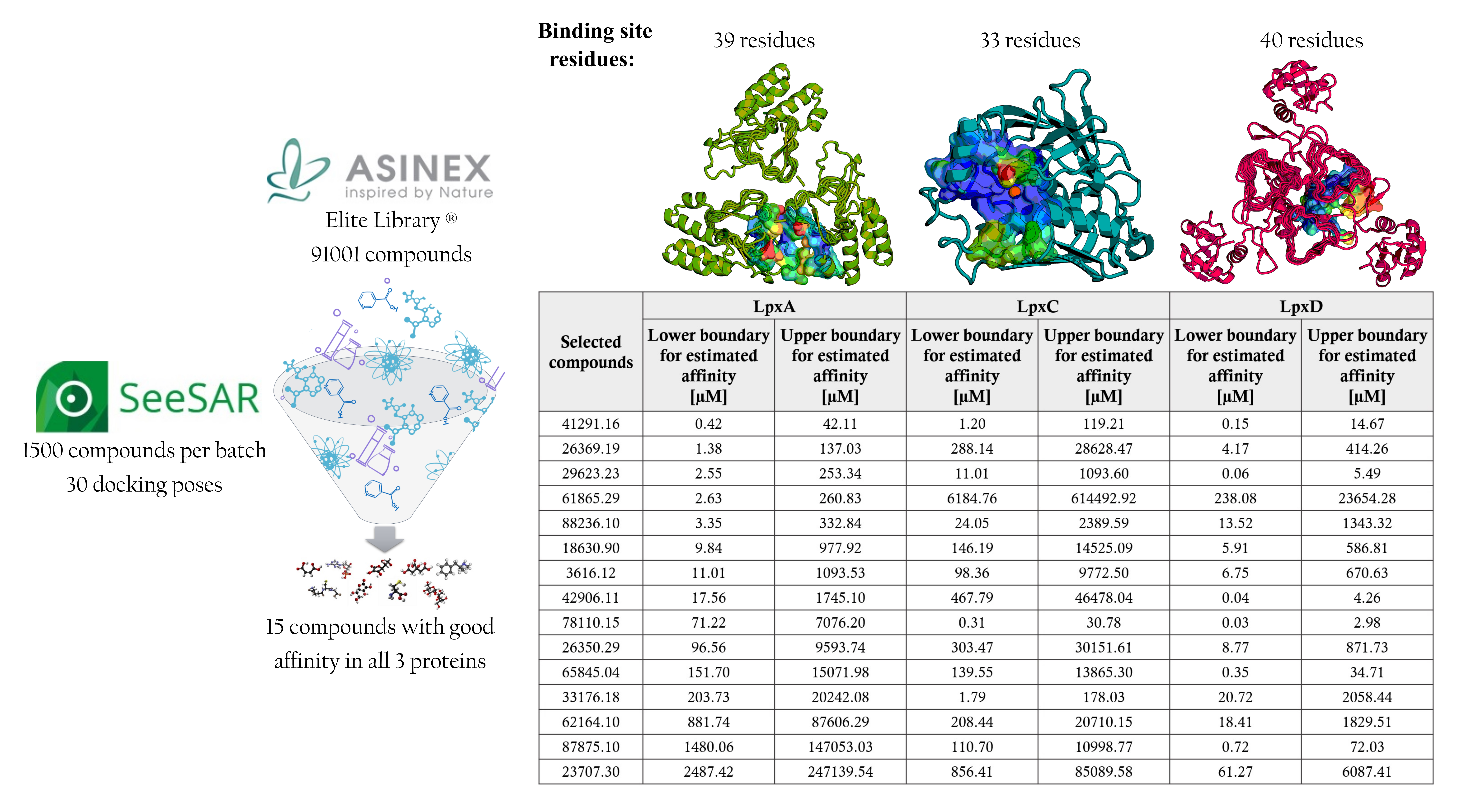
The proposed project aims to design a small molecule compound that targets LpxA, LpxC, and LpxD, the three essential acyltransferases in the lipid biosynthetic pathway which are the potential antibiotic targets for nosocomial infections and combating multi-drug resistance.
Obstacles faced:
• Screening the whole library of compounds at a single go was not possible. SeeSAR is capable of generating only 49,999 poses at a time. Hence screening a large volume of compounds was difficult and consumed a considerable amount of time.
Solution:
• The whole library of compounds was split into batches of 1600 compounds each. (1600 compounds * 30 poses = 48,000). Thus, the compounds were screened in 57 batches for each protein.
Proposed future work:
The top compounds have some bad torsions, inter and intramolecular clashes. In future, the work aims to optimize and improve the potential lead compounds obtained by the initial screening using the HYDE scoring function.
After 3 months, Soundarya has achieved the following milestones:
- In order to avoid drug failure due to lack of efficacy and safety, a screening library (Elite Library®) with 91001 lead-like compounds was retrieved from Asinex database which was already pre-screened and validated against the early ADMET panels to ensure that the drug candidates have sufficient efficacy and appropriate ADMET properties. SeeSAR was used for the initial screening of the library. The binding pockets of LpxA, LpxC and LpxD were identified using the DoGSiteScorer tool. The screening library was pre-processed to generate poses that fit into the predicted binding pocket of the proteins. A maximum of 30 poses with standard clash tolerance were used as pose generation parameters for all 3 proteins. H-bond network, clashes, torsion quality, Optibrium properties, and estimated affinity were calculated for all generated. The results were sorted using the estimated affinity values. The top 15 compounds that had a good affinity in all three proteins are chosen for further analysis.
- Yet to be done
- Yet to be done