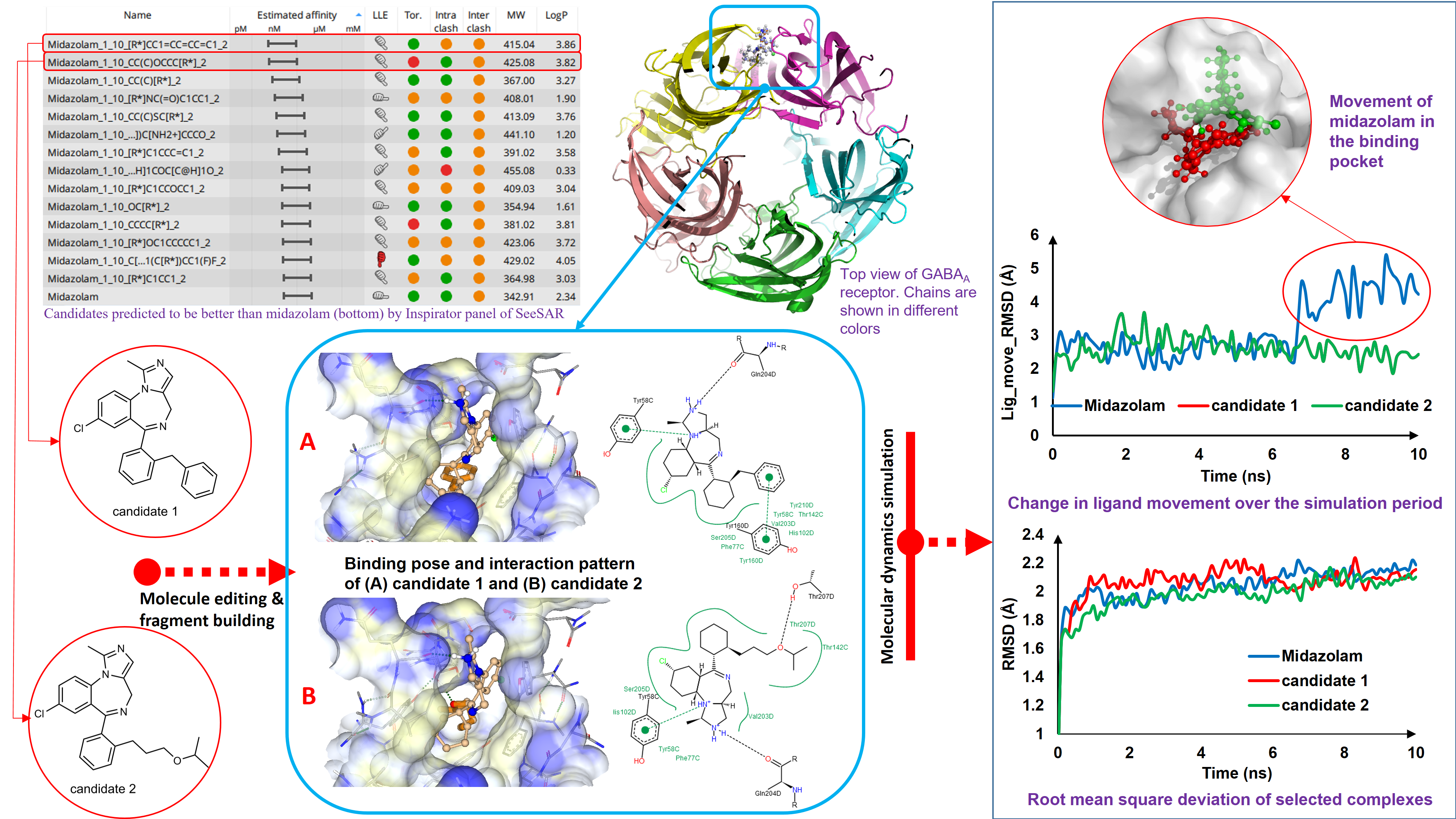
In the second phase of our study, we have edited the structure of midazolam with an attempt to improve its binding affinity towards GABAa receptor. For that purpose, fluorination at potential sites of midazolam molecule were made which yielded 54 molecules. No molecule within this set surpassed midazolam with respect to binding affinity. However, MZCF11 was the best among the modified analogues. In the next step, molecule editor panel of SeeSAR 9.1 was utilized to modify different parts of MZCF11 to improve its affinity which produced two compounds with binding strength almost equal to midazolam. Nonetheless, these compounds suffered from unacceptable intra- and inter- molecular clashes. Midazolam itself was modified using the molecule editor which yielded no fruitful results. In order to get a better candidate than midazolam, the unoccupied space in the binding pocket was analyzed. From the pocket analysis, it turned out that the phenyl fragment and the fluorine atom present in the phenyl ring could be allowed to regrow using the inspirator panel of SeeSAR 9.1. Such regrowth yielded 14 compounds better than midazolam. Best two of these, in terms of binding affinity, were again subjected to docking with an aspiration to get a better pose in terms of torsional and clash score. However, no better pose was obtained after docking. In the first phase of our study, we have identified that C:PHE77, C:TYR58 and D:TYR210 residues were critical for binding with GABAa receptor. Our best two candidates were also found to interact with these residues along with other residues indicating that these molecules followed a similar trend in binding with GABAa receptor.
In the third phase, we have subjected three drug – receptor complexes to all – atom molecular dynamics simulation using YASARA software where membrane simulation protocols were followed since GABAa receptor is a transmembrane protein. Midazolam was considered as the control and the rest two molecules were our best candidates as identified from binding affinity analysis previously. MD simulation analysis unveiled a quite stable binding of the drug candidates chosen. Interaction analysis of MD trajectory of both candidate 1 and candidate 2 revealed that they maintained a steady interaction with C:PHE77, C:TYR58 and D:TYR210 residues through hydrophobic interactions mainly throughout the simulation period. To summarize, the dynamic behavior of our selected hits revealed a stable binding pattern inside the binding cavity. Our chosen candidates were predicted to exhibit satisfactory pharmacokinetic properties by OPTIBRIUM.
We conclude that these candidates were found be better than conventional, marketed benzodiazepines by our study.
After 1 year, Surid Mohammad has achieved the following goals:
- To find out the key amino acid residues of GABAa receptor mediating its interaction with benzodiazepines and to recognize the marketed benzodiazepine showing best binding affinity.
- To fluorinate the benzodiazepine with best binding affinity (found from 1st phase of the study) to generate fluorinated analogues and test their binding strength against GABAa receptor to obtain a better candidate than the marketed benzodiazepines.
- To evaluate the dynamic behavior and binding profile of selected hits using molecular dynamics simulation.