The discovery of bioactive small molecules is an expensive and time consuming yet central task in drug discovery. A potentially superior way to identify new drug candidates is the selective optimization of side activities (SOSA), which employs known drugs for their side-activities as lead compounds. Virtually every small molecule drug interacts with more than a single molecular target and, thus, has side-activities. Sometimes, these side-activities may be of therapeutic value and structural optimization to turn the side-activity into the main activity can generate a new drug. As a key advantage of this strategy, analogues of approved drugs inherit their previously optimized favourable characteristics concerning toxicity, solubility, bioavailability and metabolic stability, and are drug-like by definition. We intended to apply the SOSA strategy to the CysLT1 inhibitor cinalukast for which we have observed a previously unknown side-activity on the peroxisome-proliferator activated receptors (PPAR) alpha and gamma. These ligand-activated transcription factors play a crucial role in metabolic disorders. To speed up the SOSA-focussed structural variation of cinalukast towards potent PPAR modulators, we intended to employ in silico techniques to support the SOSA concept.
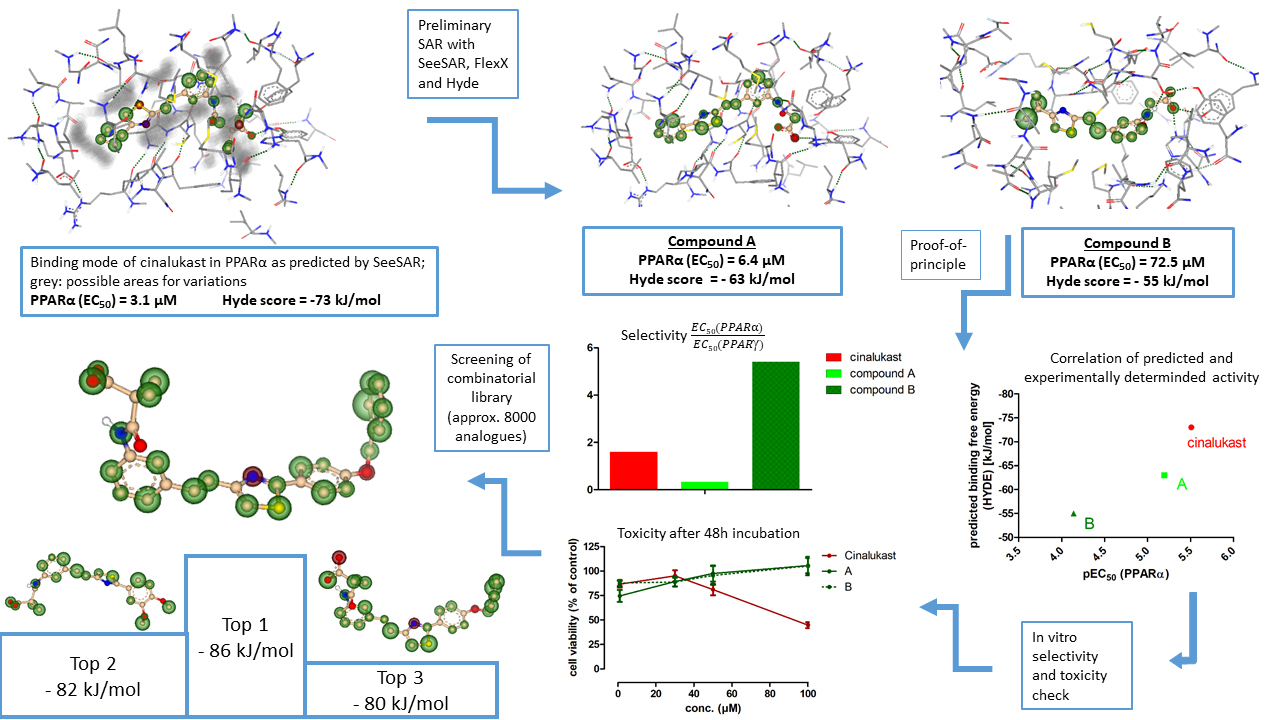
We started this endeavour using SeeSAR and its editing mode to visualize and identify key ligand-receptor interactions. Moreover, FlexX and Hyde served to analyse the binding mode of a small combinatorial set of cinalukast derivatives and allowed an in silico estimation of its structure-activity relationship (SAR). This enabled us to select structural elements whose variation promised to improve potency on PPARs. As a proof-of-principle, two cinalukast analogues were prepared and tested in vitro. Their experimentally determined modulatory activity on PPARs nicely correlated with the predictions of Hyde confirming the suitability of our computational approach to support SOSA. Moreover, these cinalukast analogues revealed remarkable improvements in toxicity over the lead compound.
Next, we generated a combinatorial library of approx. 8000 cinalukast derivatives using adaptions of the KNIME workflows STORM and MedChemWizard. The BioSolveIT KNIME nodes were then employed for automated docking and screening to identify the most promising candidate compounds in the library.
With the implementation of this automated and time-saving workflow for a computationally guided SOSA approach, we are speeding up the structural optimization of cinalukast towards potent PPAR modulators and several predicted analogues are in preparation and characterization. This successful combination of computational tools in compound optimization highlights the potential of computer-assisted SOSA for future drug discovery.
After 1 year, Julius has achieved the following goals:
- First, we intended to identify variations of our lead structure cinalukast to counter the compound’s high toxicity and improve key characteristics such as selectivity or solubility. We employed SeeSAR to study a manually defined library of 40 analogues comprising variations which we chose as a starting SAR. We observed that the core scaffold of cinalukast provides an ideal angle for the ligand-receptor interaction and therefore our initially intended changes in this part were discarded. Thereby, the computational approach significantly reduced the number of analogues that had to be prepared. The most promising analogues according to SeeSAR were further investigated with FlexX and Hyde scoring to analyse ligand-target interactions and binding contributions. Two analogues were selected for synthesis and in vitro characterization taking into account all computational results to confirm a correlation between predicted and experimentally determined values.
- As a second goal, we aimed for broader variations of the lead structure to improve potency on the new target. We tried to employ the KNIME workflows MedChemWizard and STORM and successfully adapted them to our needs. With the basic concept of these workflows and using KNIME nodes provided by BioSolveIT, we built and screened a large combinatorial library of approx. 8000 analogues of the lead compound. Predicted information on compound binding obtained from SeeSAR helped the library design. After the automated library generation and screening workflow, we manually analysed top-ranking compounds by docking with the SeeSAR and using the “Interactive BioSolveIT table” for various features. The three most potent derivatives from the automated screening workflow turned out very favourable and were selected for synthesis and in vitro characterization.
- As a third objective, we aimed to confirm the predicted data (goal 1 and 2) in vitro by synthesizing and testing the selected analogues. To confirm the suitability of our computational approach, we selected two compounds for synthesis to study the correlation between predicted and experimentally determined value. The in vitro activities of these analogues nicely correlated with their predicted Hyde scores thus characterizing this ranking approach as suitable for our optimization strategy. Moreover, we selected compounds for synthesis to reduce the problematic toxicity of the compound class which was achieved by minor structural changes deduced from SeeSAR calculations. Finally, we employed the previously validated Hyde scoring as decision method for selecting compounds from a large combinatorial library to optimize potency of the compound class with as few analogues to be prepared as possible and the three top-ranking candidates are in synthesis/in vitro characterization.