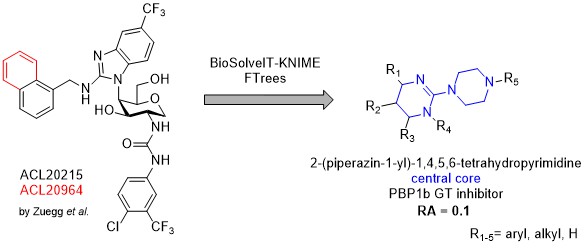
Peptidoglycan transglycosylases or alternatively called glycosyltransferases (GTs) are membrane-bound enzymes that catalyze the polymerization step in cell-wall biosynthesis. They represent a prominent target in antibacterial drug discovery and only a few inhibitors are described in the literature. Moreover, native substrates Lipid II/Lipid IV display considerable structural complexity while previously described inhibitors often closely follow their complex structural motifs. Moenomycins for example, are structured as a tetrasaccharides with an isoprenoid chain connected through a phosphoglyceric acid moiety. Recently, monosaccharide disubstituted with two aromatic moieties, comparatively smaller, inhibitos of glycosyltransferases, have been described by Zuegg et al. Theese compounds could facilitate development of novel small molecule GT inhibitors. As the binding mode of described inhibitors is not known, four ligand based drug design campaigns have been performed with the help of BioSolveIT KNIME tools ammounting to the library of 83 purchased compounds that were all biologically evaluated for their relative inhibition of the transglycosilase reaction of PBP1b from E. coli using a fluorescence assay. Three structurally similar hits were obtained where one compound displayed residual activity of PBP1b at 10 % and is expected to possess IC50 in the low micromolar range. In order to unequivocally describe the PBP1b inhibition, further Lipid II preparation is under way for the support of biological testing and a synthetic campaign has been started to resynthesize purchased hits with preparation of relevant analogues, especially lipophilic-chain conjugates. Latter compounds could shed further light on Lipid II mimicry if present and be used as molecular probes for further inhibitor design/optimisation.
After 1 year, Marko has achieved the following goals:
- First and most important phase 1 goal (all phase 1 goals accomplished). Establish and set up bological evaluation support. From the start of campaign, biological evaluation support has been established where purchased compounds can be evaluated for PBP1b inhibition with a fluorescence test. Substrate consumption by PBP1b can be monitored through fluorescamine labeled Lipid II and HPLC analysis. Test system consists of E. coli enzyme, Lipid II substrate in a suitable buffer/detergent system. Furthermore, in vitro antimicrobial testing and cytotoxicity testing using human embryonic kidney [HEK] 293 cells awaits for (re-)synthesized hit compounds and analogues. Here the emphasis was on the complexity of test system and availability of Lipid II substrate.
- Milestone 1 (phase 2) – Identify a hit compound (or compounds) on the basis of 1st, 2nd or multiple iterations of ligand based drug design. With the help of BioSolveIT KNIME tools, 4 iterations of ligand based drug design have been performed. Final library of purchased compounds consists of 83 evaluated compounds and three identified hits. Hit compounds are all structurally related and bear a central 2-(piperazin-1-yl)-1,4,5,6-tetrahydropyrimidine moiety. We hope we will be able to prepare suitable structurally decorated analogues that would display even greater activity on isolated PBP1b and maybe even posess antimicrobial activity. Both outcomes are favourable as molecular probes for the study of glycosyltransferase inhibition are more than welcome.
- Milestone 2 (phase 2) – Perform further in silico studies extending from binding mode studies to supporting in silico studies in hit identification and design. With the KNIME – BioSolveIT synergy, multiple workflows have been developed that span from library filtering, PAINS identification, similarity search and visualisation of results. First three ligand based design iterations were focused on monosaccharide disubstituted query molecule, while the fourth campaign has been tailored to expand the chemical space of a first hit compound identified. With the fourth ligand based design iteration, we identified additional two hits with further enhanced activity on E. coli PBP1b. Following resynthesis of hits and relevant analogues, additional structure-based studies will be conducted using BioSolveIT-KNIME integration. The emphasis of the whole project resides on lean and transparent hypotheses in design phase where ligand based workflow is used and extensive experimental support.