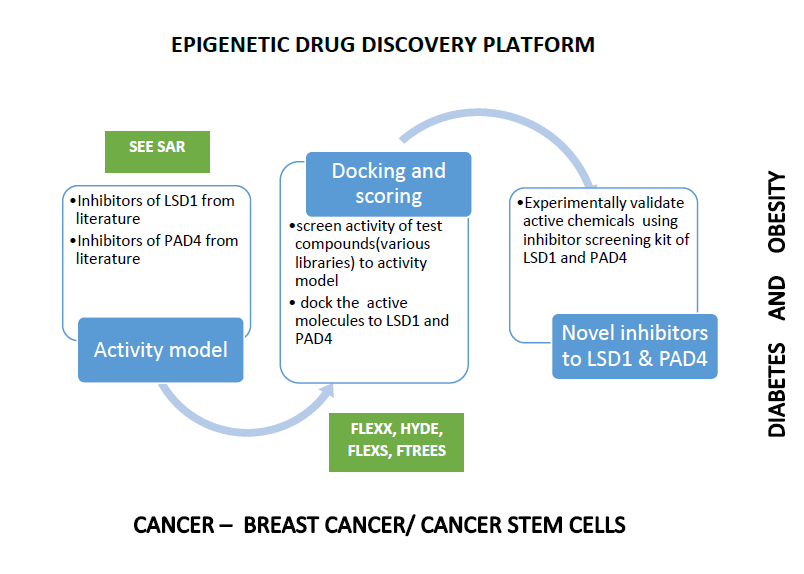
Natural products have proven to inhibit Lysine-Specific Demethylase-1 (LSD1) were retrieved from literature, and structure-activity relationship (SAR) is depicted using See SAR software. See SAR helped us to visualize hydrophobic, electrostatic and other interactions responsible for inhibition of LSD1. There were no natural products proven to inhibit Peptidyl Arginine Deiminase 4 (PADI4). We had docked manually curated natural product library retrieved from PubChem library, and four natural products were chosen to validate experimentally. We had purchased the natural products from commercial vendors and inbuilt LSD1, and PADI4 inhibitor screening assays were carried out. It resulted in universal inhibitor to inhibit both proteins LSD1 and PADI4. We had utilized ReCore module from LeadIT to develop the derivatives of common inhibitors for the potent inhibitor. We utilized FlexX docking tool to predict the binding affinity and estimated the Gibbs free energy using SeeSAR and as well FlexX score. We repeated the process till we identified inhibitors possesing free energy -kCal/mol. Use of See SAR, FLexX, and ReCore modules resulted in best scoring leads to inhibit PADI4 and LSD1
After 1 year, Veera Chandra Sekhar Reddy has achieved the following goals:
- The primary objective of goal 1 was to developed Structure-Activity Relationship (SAR) on natural products to inhibit LSD1 and PADI4 based on literature using SeeSAR and FLexX docking methodology. Proven natural products to inhibit LSD1 are resveratrol, Curcumin, quercetin, myricetin, luteolin, apigenin, genistein, and TCP. The respective three-dimensional structures(natural products) was retrieved from PubChem, and the poses were calculated using FlexX score and the same are used to predict the structure-activity relationship model. To complete the goal to build the model for PADI4, as there were no proven chemicals to inhibit PADI4, we had screened natural products from library using FlexX score and based on experimental studies new model was built on chemicals to inhibit PADI4
- The main objective of goal 2 is to perform flexible docking and chose the best scoring chemical based on the rank, score and interacting residues. This goal is to address the optimization of leads to LSD1 and PADI4. ReCore module is used to design, develop derivatives of top scoring leads to inhibit our drug targets. Briefly, natural product library was retrieved from National Cancer Institute (NCI) and the library is used in the process of flexible Docking. FlexX tool is used to perform protein-ligand interactions using Flexx score and the top scoring natural products were studied for their binding interactions The top ranking pose is used as input for ReCore module to develop derivatives. having visual on binding pose view, interacting residues and the scope of expanding the natural product had helped us to develop derivatives.
- The main objective of goal 3 is to validate initial leads also called as hits to validate experimentally and use the same tools to optimize the proposed leads. Five natural products were purchased from commercial vendors, and they are validated experimentally using inbuilt experimental assays. SeeSAR, FlexX and ReCore modules were used to develop their derivatives, and a library of 100 chemicals was designed and flexX is used to score the derivatives. We are in the process of synthesis and validating the new derivatives to inhibit LSD1 and PADI4.