Epidermal growth factor receptor is one of the most studied cancer drug target of pharmaceutical relevance. Several first generation inhibitors of EGFR (Gefitinib and Erlotinib) are in clinical use targeting L858R mutant EGFR. However, acquisition of a secondary point mutation (T790M) leads to acquired resistance to these inhibitors, rendering gefitinib and erlotinib ineffective. Consequently, the search for mutant specific inhibitors led to identification of few covalent molecules which have shown limited clinical efficacy and toxicity. Recently, due to toxicity issues of covalent inhibitors, there has been a shift in the focus of the researchers which is now inclined to identify reversible inhibitors which target both EGFR activating mutation (L858R) as well as drug resistant mutant (T790M) i.e. TMLR inhibitors. Thus, in this project we proposed to identify reversible inhibitors against TMLR mutant EGFR, using ligand and structure based drug design approach via tools available in BioSolveIT software package.
Although it is well known that a hit molecule must not only have high affinity against the drug target but also possess good ADME properties yet most drug discovery programs show high attrition rate during early phase of drug development. The reason being that most of the inhibitor identification programs concentrate on identifying highly active ligands but neglect other parameters which make these ligands unattractive for drug development. Therefore, it is important that drug discovery studies are not only based on affinity and potency alone but also aims at identifying molecules which have optimized ADME properties. Thus during the entire period of the Scientific Challenge our aim was to identify/design a few potential new ligands/hits which have not only high affinity against EGFR-TMLR mutant but also favorable ADME properties like low MW, LLE, LE, logP and TPSA.
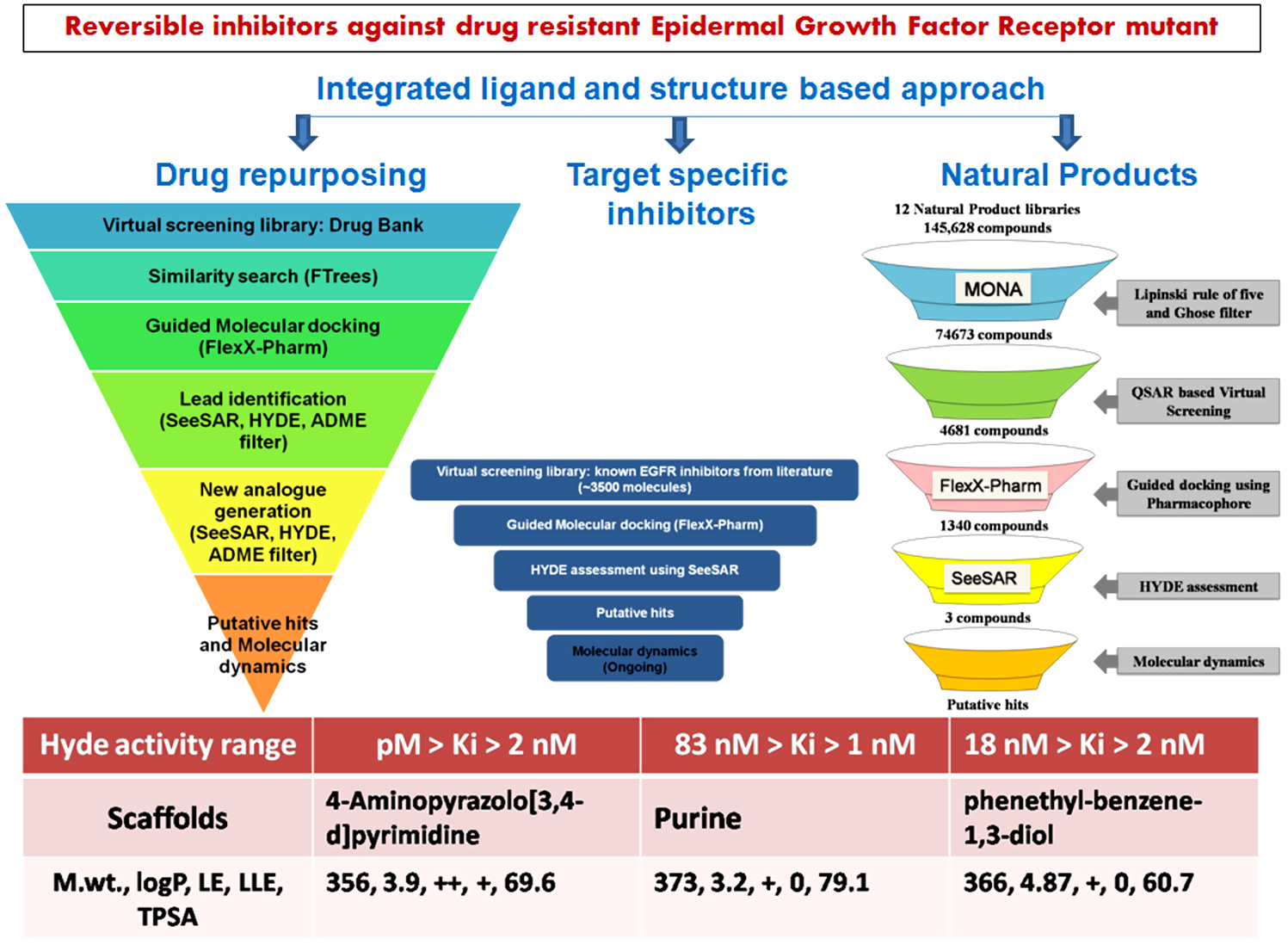
As hits that serve as starting points for lead discovery are invariably made available from either of the 3 resources i.e. (i) known drug like compounds i.e drug repurposing (ii) natural products and (iii) scientific literature; we had designed the computational experiments in a manner which allows identification of putative hits from each of these respective approaches enabling detection of various scaffolds that could later be optimized by experimentalist.
The range of modules available in BioSolveIT software including (FlexX-Pharm, FTree, MONA, SeeSAR, HYDE and ADME prediction) along with integration of different approaches (ligand, structure, QSAR) have enabled us immensely to achieve the below mentioned goals.
After 1 year, Dr. Subhash Mohan Agarwal And Dr Prajwal Nandekar has achieved the following goals:
- Discovery of TMLR inhibitors by drug repurposing For discovering structurally diverse novel compounds from DrugBank we used BiosolveIT's FTrees, which searches for similar molecules against the query. We then used FlexX-Pharm to screen the constructed library followed by HYDE assessment in SeeSAR. It allowed us to identify a few low molecular weight scaffolds which occupy the critical sub-pockets and have desired interaction. Using one of the scaffold (MW= 295, LE +, LLE 0, logP 3.1) in SeeSAR, we then evolved new molecules having higher interaction affinity and desired ADME properties. The designed compounds have favorable LLE (+), LE (++), low MW (<400), logP (3.9-4.7) and TPSA (~70) and low nM predicted activity as indicated in SeeSAR. Also, we undertook molecular dynamics simulations to check stability of ligand in active site. Thus, we employed integrated ligand and structure based approach using BiosolveIT for identifying inhibitors from known drug molecules.
- Screening of natural product libraries for identification of TMLR inhibitors We have screened 12 NP-databases for their oral bioavailability and drug-likeliness using Lipinski and Ghoose filter available in MONA. We then predict the anticancer activity using a published QSAR model recently developed by me. We next examined interactions of these molecules with the EGFR-TMLR protein through guided molecular docking using FLexX-Pharm. The docking results were then imported in SeeSAR for HYDE assessment & visual analysis. We identified a few molecules that have low molecular weight and favorable LLE, LE, logP and TPSA as well as low nM predicted activity as indicated by HYDE affinity parameter. We also evaluated the stability of interactions using MD. Thus in the present work we have integrated MONA based physiochemical filtering, QSAR approach, FlexX-Pharm based molecular docking, SeeSAR hyde assessment and MD for identifying inhibitors against mutant TMLR protein from NP-libraries.
- Exploring EGFR targeted synthetic library for prioritizing TMLR inhibitors We have used literature curated synthetic inhibitors of EGFR taken from our published central repository EGFRIndb for virtual screening. We selected the library as it contains inhibitors that act against the wild isoform along with IC50 values, are sure about their synthetic feasibility and can be used as reference for understanding selectivity. We have screened these inhibitors using FLexX-Pharm followed by HYDE assessment in SeeSAR. Our initial result indicate 3 molecules have low nM binding affinity however, the molecules do not comply with all the desired ADME properties (MW, LLE, LE, logP and TPSA).