Prion disorders are fatal neurodegenerative conditions caused by the conformational conversion of the normal, cellular prion protein (PrPC) into a misfolded isoform (PrPSc) that accumulates in the brain of affected individuals. A possible strategy for tackling prions is to stabilize the monomeric native conformation of PrPC, thus inhibiting its misfolding. This goal could be achieved with PrPC ligands capable of acting as pharmacological chaperones. Interestingly, the impact of such ligands would not be limited to prion diseases. In fact, recent evidence suggests that PrPC could transduce the toxic effects of a variety of misfolded proteins, such as oligomers of the amyloid β peptide (Aβ), and other disease-associated aggregates. Therefore, the identification of PrPC binders might possibly be relevant for several neurodegenerative conditions.
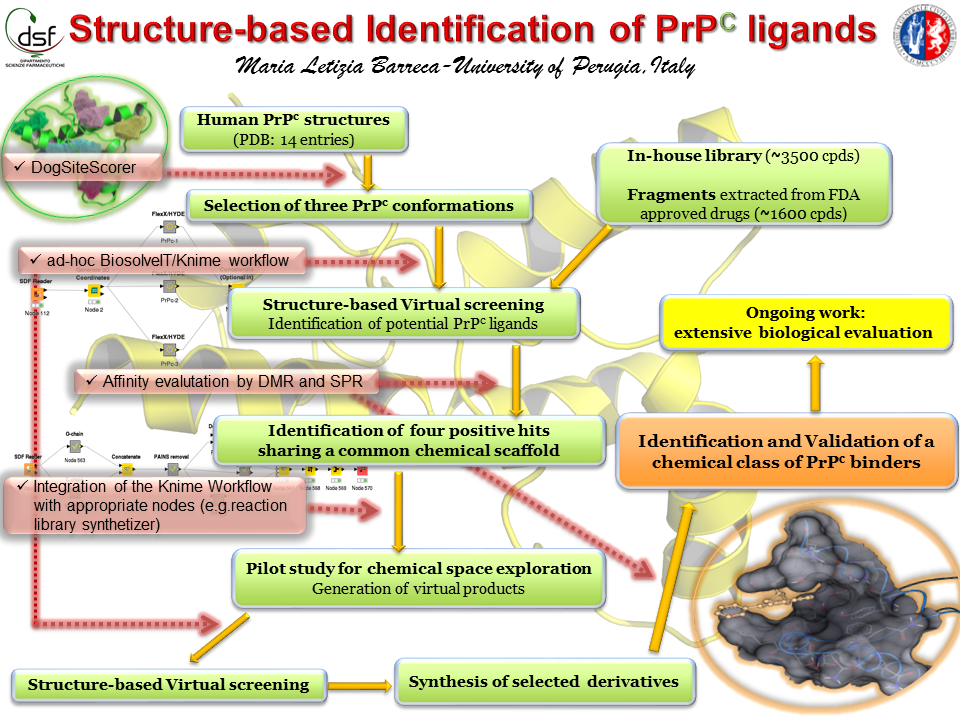
The main goal of this project was to rationally identify PrPC ligands with the help of BioSolveIT softwares. Available conformations of human PrPC were analyzed by DoGSiteScorer for protein druggability assessment. The analysis suggested three PrPC protein conformations as the best targets for virtual screenings. At this point, we found really helpful the possibility to integrate BioSolveIT tools into KNIME. In fact, to assist the achievement of our goal we built an ad-hoc KNIME workflow for the identification of potential PrPC ligands.
Two different libraries (fragments and in-house molecules) were thus screened against the three protein conformations, and 50 potential PrPC binders were selected for experimental evaluation. Next, we employed two complementary biophysical techniques (i.e. dynamic mass redistribution, DMR; and surface plasmon resonance, SPR) to characterize the binding of the virtual hits to PrPC. Five compounds were confirmed as PrPC ligands with affinity constant (Kd) ranging from 50 to 500 microM; of note, four of them belong to the same chemical family and showed high IP-value.
The encouraging finding prompted us to focus our efforts on the identified chemical scaffold and to use the potentiality of CoLibri tool to design a focused set of new compounds structurally related to the hits. The chemical space exploration approach was integrated into the previously developed KNIME workflow and a pilot study was performed by using the in-house chemistry and our own building blocks. Among the in silico generated derivatives suggested by FlexX/HYDE as potentially able to bind PrPC with stronger affinity compared to the parent compounds, the four simplest structures were selected for chemical synthesis. The synthesized derivatives were evaluated for their binding to PrPC by DMR and SPR, and two improved PrPC ligands (MW around 380 Da) were identified and are now under extensive biological evaluation.
After 1 year, Maria Letizia has achieved the following goals:
- Development of an ad-hoc BioSolveIT/KNIME workflow to identify potential PrPC ligands. Fourteen protein conformations of human PrPC were retrieved from the RCSB PDB and processed by DogSiteScorer to assess protein druggability. Three PrPC conformations were thus selected as targets for structure-based virtual screening. We have then designed, built and configured an ad-hoc KNIME workflow, which included 1) Receptor preparation and virtual screening of two compound libraries using LeadIT; 2) Assessment of the ligand affinity with HYDE in SeeSAR; 3) Filtering out of compounds with low estimated affinity, unfavorable predicted value of LE and classified as “-“ in the BBB category. The results obtained for each target were concatenated in such a way to easily identify potential PrPC binders conserved across the three different conformations. Next, visual analysis and chemical structure clustering allowed us to identify 52 virtual hits for experimental validation.
- Identification of a new chemical class of PrPc ligands. In order to evaluate the binding of the in silico selected compounds to PrPC, we developed a novel screening method based on DMR, a label-free, fully automated biophysical technique capable of detecting molecular interactions at the equilibrium. Positive hits were then further characterized by SPR, a complementary biophysical technique. Five virtual hits were experimentally confirmed as PrPC binders by both techniques (Kd range: 50-500 microM). Notably, four of such hits were in-house developed compounds sharing a common chemical scaffold and possessing high IP-value.
- Successful pilot study for chemical space exploration. The binding affinity data prompted us to conduct a pilot study where a restricted chemical space around the identified PrPC ligands was designed and built. For this purpose, the in-house developed KNIME workflow was integrated with appropriate nodes (e.g. Reaction Library synthesizer). The chemical reactions were defined based on our chemistry experience, and in order to speed up the synthesis of possible virtual products, only reagents available in our labs were used as building blocks. Thousands of virtual products were generated and then submitted to virtual screening. HYDE predicted several derivatives as higher-affinity PrPC binders compared to the starting hits. In order to validate the approach, the four simplest structures were synthesized and experimentally evaluated. Encouragingly, two derivatives showed improved binding profile, and their extensive biological evaluation using assays for prion propagation is undergoing.