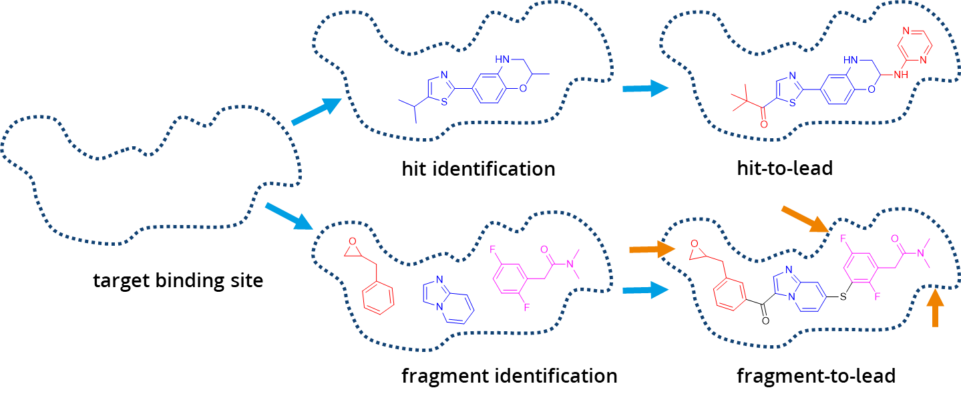
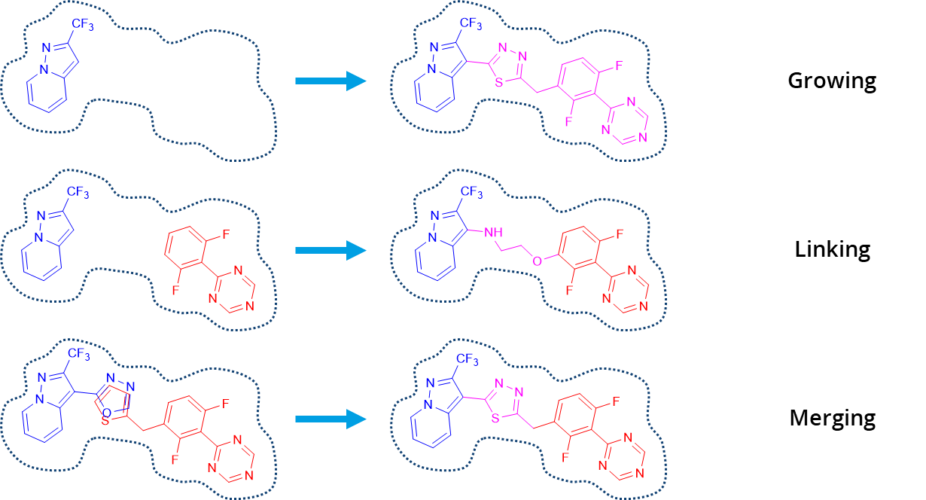
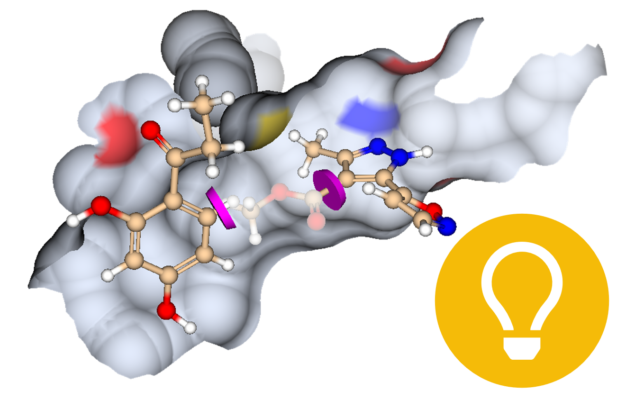
All of the mentioned methods can be applied in SeeSAR. In the Inspirator Mode it is possible to perform fragment growing, linking, and merging. The library for growing can be easily modified by users to create a custom version with their own building blocks. Furthermore, users can explore the binding pocket by themselves within the Molecule Editor Mode and test their hypothesis with the visual feedback they receive from SeeSAR.
Several examples of FBDD with SeeSAR are available as tutorials.