SeeSAR 13.1 is here and it comes with a truckload of new features! The update to ‘Midas’ is not only able to synergize with the new BioSolveIT platform HPSee, but will convince with a plethora of quality-of-life improvements.
Therefore it may not surprise, that we are especially proud of the covalent docking augmentation. With SeeSAR 13.1, covalent docking becomes as easy as ABC; Covalent warheads are now automatically recognized during docking runs. Pre-processing of ligands or compound libraries is therefore no longer required. Simply select a residue in the binding site that you would like to target for the covalent docking and start the pose generation. With this update, SeeSAR automatically detects and transforms 36 most commonly used covalent warheads during a covalent docking run.
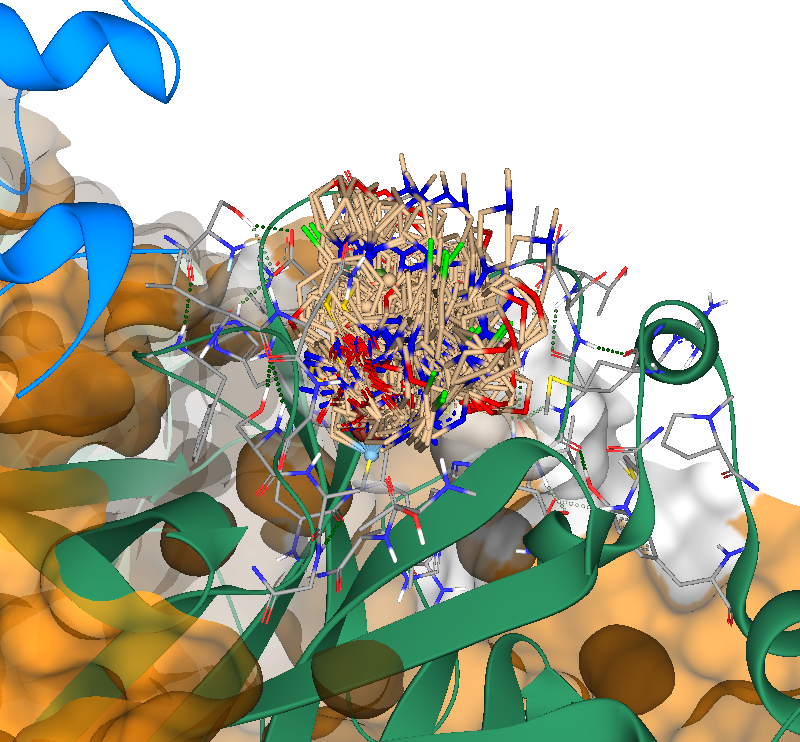
Results of a docking run on PDB 7TLL. With only a few clicks, 20 molecules containing a covalent warhead were docked within seconds.
Seasoned SeeSAR users can still perform the old docking procedure by introducing [R] linkers to their molecules for covalent docking.
You should absolutely download and test this feature. Covalent docking has never been more satisfying.
Another huge spotlight should be put on the extension of the Docking Mode. Now, coupled with the new workflow environment HPSee, it is possible to conviently AND efficiently perform large-scale virtual screening campaigns on external hardware. Coupled with SeeSAR’s intuitive user interface, it becomes a pleasure to run campaigns and browsing through millions of molecules in a flash.
Of course there are a lot more things that made it into 13.1. You can read about the full set of additions and improvements in the elaborate SeeSAR changelog. But before you do that; Why not downloading SeeSAR 13.1 following this link first?





