Vast Chemical Space Navigation: We are thrilled to share the launch of infiniSee ‘Artemis’ 5.1, which comes with numerous innovative enhancements designed to enhance the compound selection process. Among the standout additions in this update is a sleek Bemis-Murcko scaffold clustering within infiniSee’s Analyzer Mode.
Introducing Bemis-Murcko Clustering
A key element driving the compound selection process is the selection of a chemically diverse set of molecules to increase the chances of finding actives. Diversity within a set offers the added benefit that, ideally, multiple individual molecular scaffolds are discovered, which can serve as potential starting points for an optimization series.
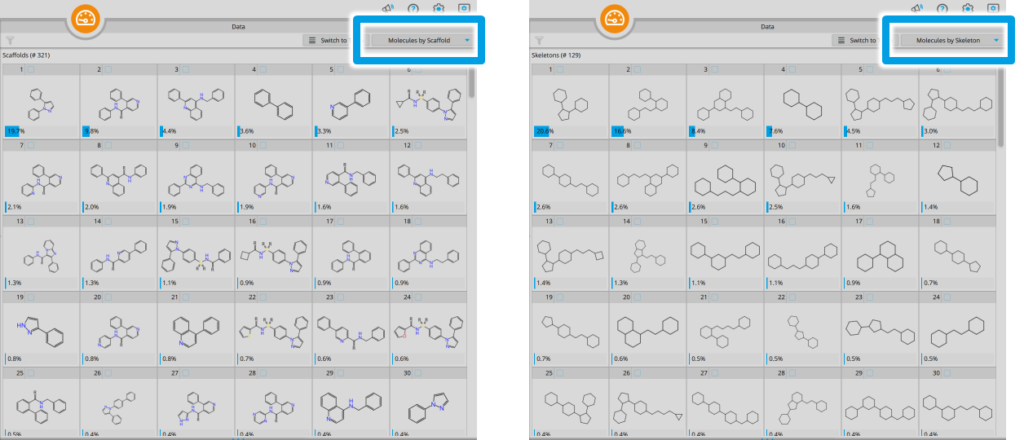
The novel compound clustering feature in the Analyzer Mode: Create groups of molecules based on their Bemis-Murcko scaffold or their mere skeletons.
The much requested introduction of Bemis-Murcko scaffold clustering augments this step: Given a set of compounds in the Analyzer Mode (e.g., infiniSee results from the Scaffold Hopper or the Analog Hunter Mode, individual molecule libraries, SMILES, …), entries can be categorized by molecular scaffolds and skeletons in the data visualization window. The frequency of calculated scaffolds or skeletons is given for each group — which in turn allows you to spot unique patterns, identify commonalities, and gain valuable insights into the composition and diversity of your compound dataset.
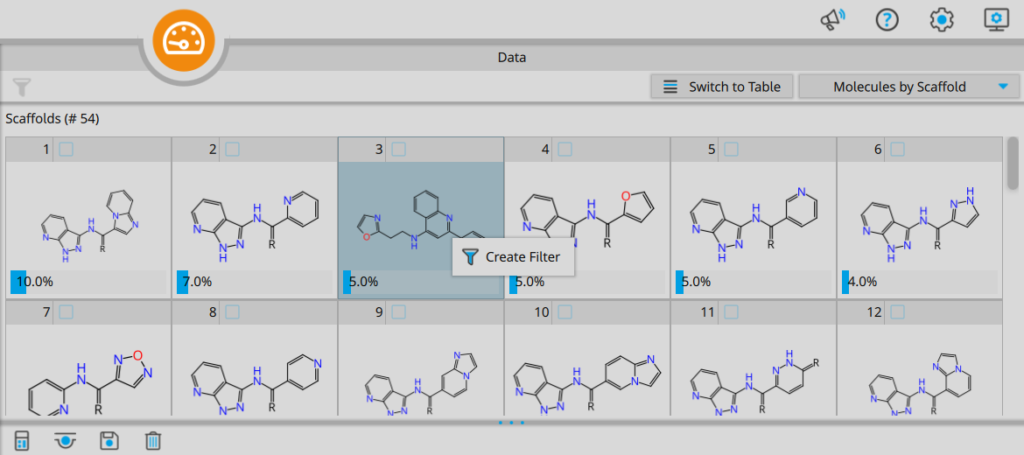
Subsets can subsequently be checked for their content to select the most interesting candidates. The filtering option allows you to browse through the comprised entries one-by-one to cherry-pick molecules from different sections of the Chemical Space.
Now, data scientists and post-processing enthusiasts perk up your ears: Processed results in infiniSee’s Analyzer Mode will be tagged with the respective molecular scaffold and skeleton with a SMILES string which can be of great interest for any form of subsequent analysis, transformations and machine learning approaches. Simply export your results in a format of your choice (sdf, SMILES, xlsx, csv) to dive deeper into your compound prioritization.
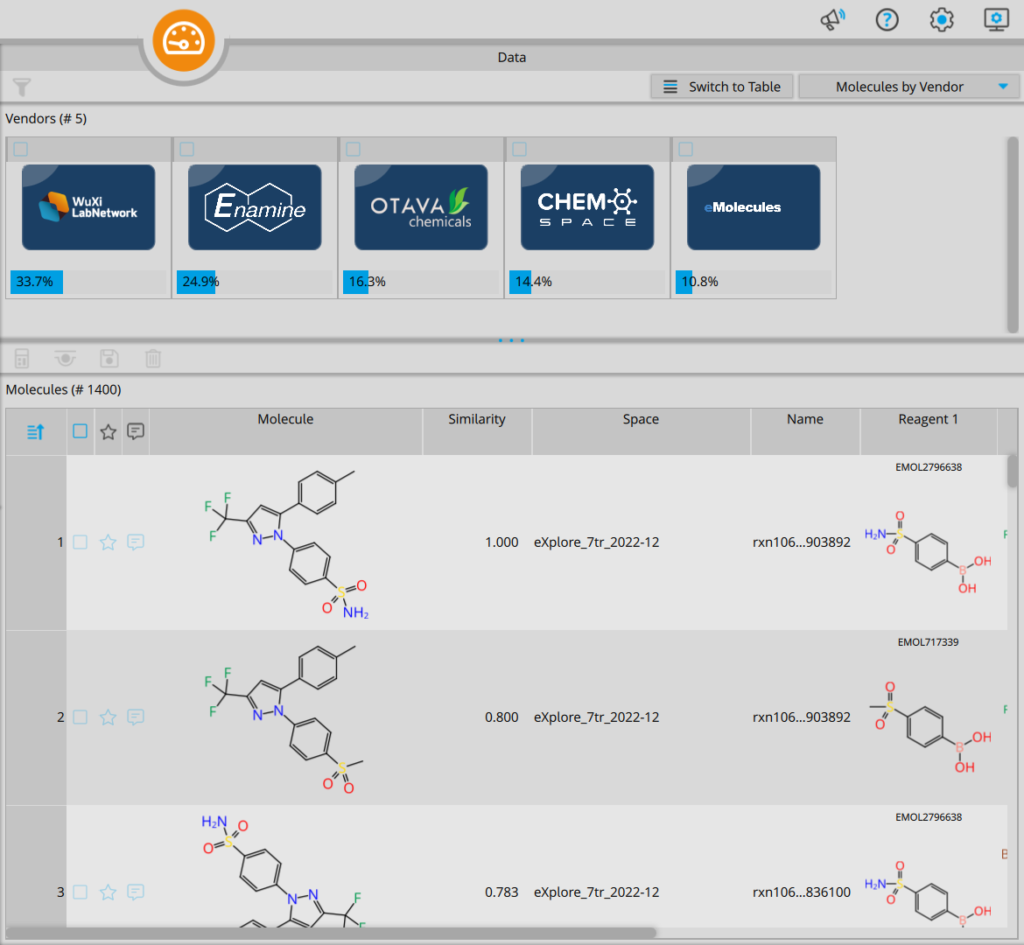
Another highly anticipated feature, fervently requested by dedicated infiniSee users, is the ability to effortlessly categorize molecules according to their respective compound vendors. This too can now be easily achieved in the data visualization window of the Analyzer Mode. In line with the scaffold and skeleton clustering, you can use the corresponding vendor Chemical Spaces as filtering criteria.
Augmented Analog Hunter Result Representation
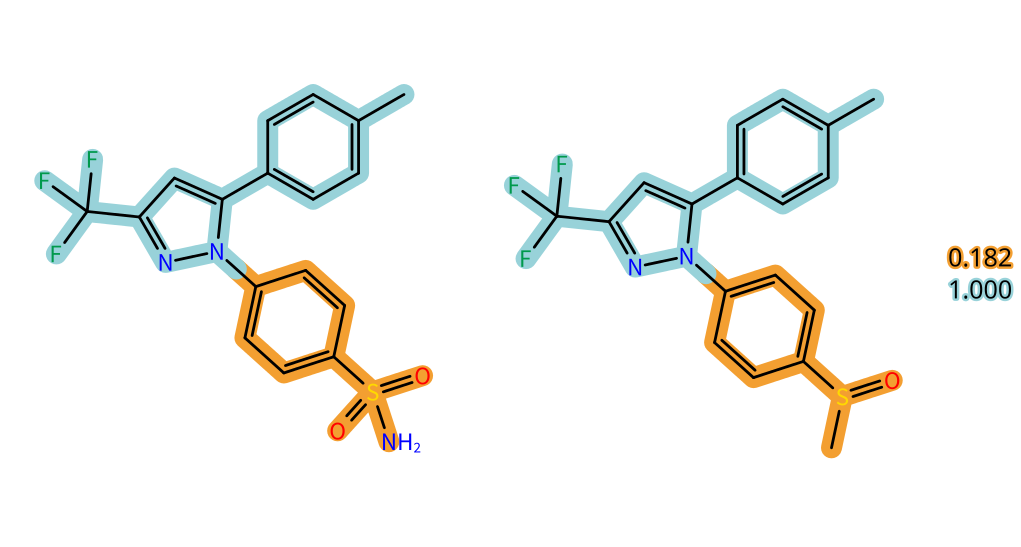
Last but not least, visual highlighting is introduced to the Analog Hunter Mode. Running the SpaceLight algorithm, Analog Hunter retrieves close analogs to a query compound from Chemical Spaces based on Tanimoto molecular fingerprint similarity.
The colorful depiction helps users to understand the fingerprint alignment of the building block substructures to the query molecule to spot areas of high similarity and differences. Additionally to the global fingerprint similarity given for each molecule, individual local similarities of the substructure matching are presented in the 2D window. Ultimately, this feature also provides hints on the building block structures used in the synthesis of the result molecule.
These innovative features work in harmony with the existing components and tools within infiniSee, offering a seamless integration that will significantly elevate the efficiency and effectiveness of the compound management and selection procedures. Give it a try and test the Chemical Space navigation platform.
The full changelog of the infiniSee 5.1 update can be found following this link.





