The covalent docking software update of SeeSAR 11.2 Hephaestus features a broad set of improvements and is accompanied by a new BioSolveIT workflow: CovXplorer provides users with a framework to conveniently explore covalent binding sites and ligands that can easily be modified to match the needs of the project.
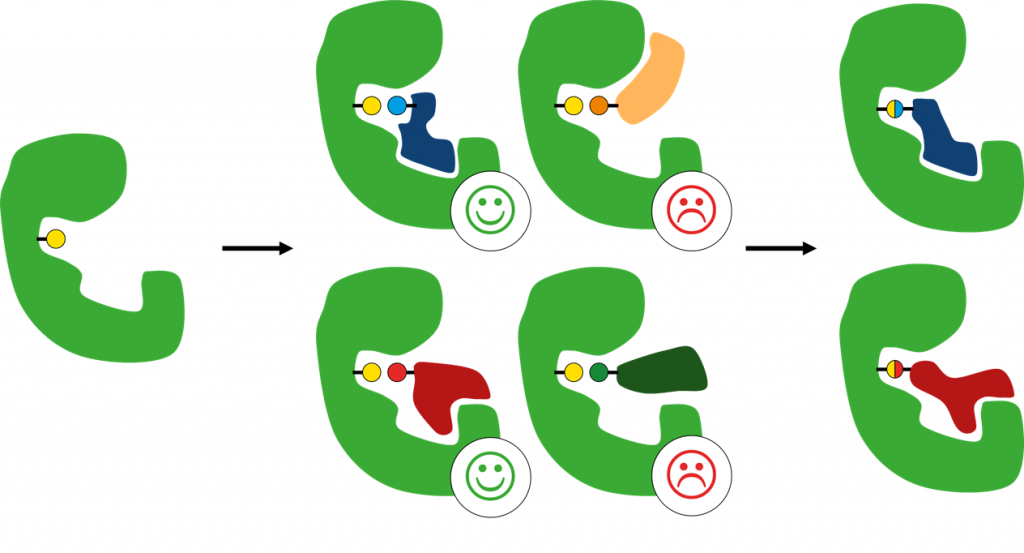
The new CovXplorer workflow mimics two stages of covalent ligand binding (see Figure 1).
In the first step the ligand is docked non-covalently in the binding site where the electrophilic warhead (e.g. a Michael acceptor) is placed in close proximity to the targeted residue. This allows an evaluation of initial complex formation before forming a covalent bond to improve the prediction of potential binders, as a strong initial reversible binding of a ligand is a good prerequisite for subsequent covalent bond formation between target and ligand. The source of the compounds can either be external compound libraries or in-house structures. After generation of docking poses they are assessed for their binding affinity, and promising candidates are transferred to the next stage. Here, warheads are processed and translated into their ligand binding form. Over 30 known covalent warheads are pre-included in the workflow and further additions can easily be added. In the final step the processed compounds are docked covalently and once again assessed for their binding affinity with Hyde to allow ranking and visual inspection of the results to select compounds for a follow-up.
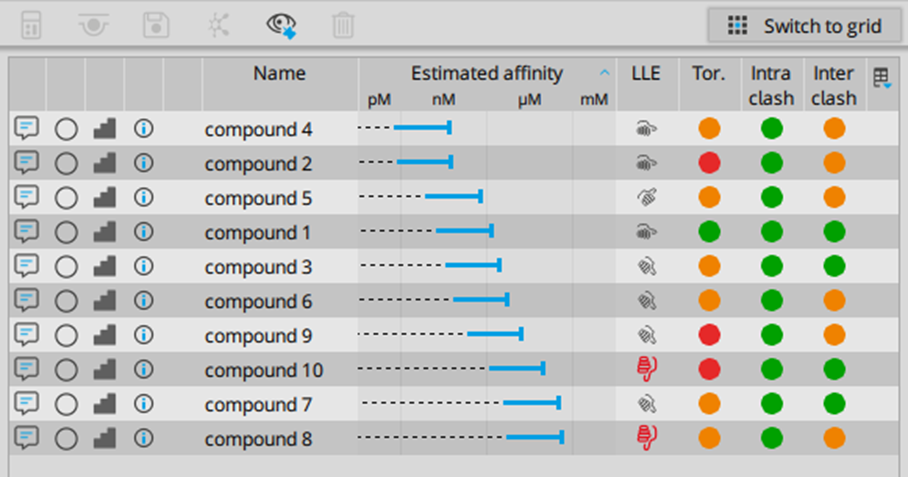
The assessment of binding modes can be visualized in SeeSAR (see Figure 2). Users profit from their knowledge and experience to dissect the generated binding modes for further data analysis. SeeSAR provides detailed and visual information on several molecular parameters of the ligand as well as on the complex itself — including molecular torsions, lipophilic and general ligand efficiencies (based on the calculated Hyde score), intra and intermolecular clashes, and particular atom interactions. Furthermore, given the design of the workflow, it is possible to filter for particular warheads of interest.
The updated binding site export as well as the improved covalent Hyde assessment provide further support to create individual solutions and workflows. With the newest version of the covalent docking software SeeSAR “Hephaestus” and the new CovXplorer users have all the tools at hand to kick-start their covalent projects and discover novel warheads and scaffolds. Profit from the accessibility of covalent vendor libraries or search in your own in-house libraries for hit candidates.
- Download SeeSAR, and start your covalent drug discovery project here.
- Get the convenient CovXplorer workflow here.



