KNIME Support
Unfortunately, KNIME is no longer actively supported by BioSolveIT.
For seasoned users, access to details on legacy workflows for older versions remains available.
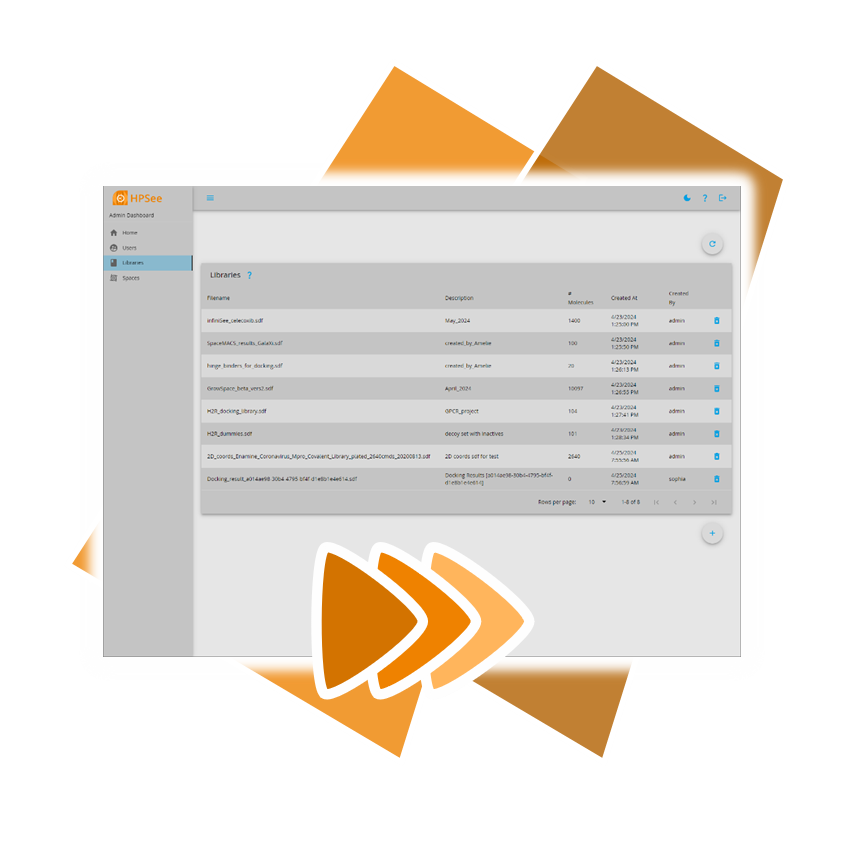
However, there's no need to rely on outdated solutions, as BioSolveIT introduces a new workflow environment tailored to the needs of computational chemists and data scientists: HPSee.