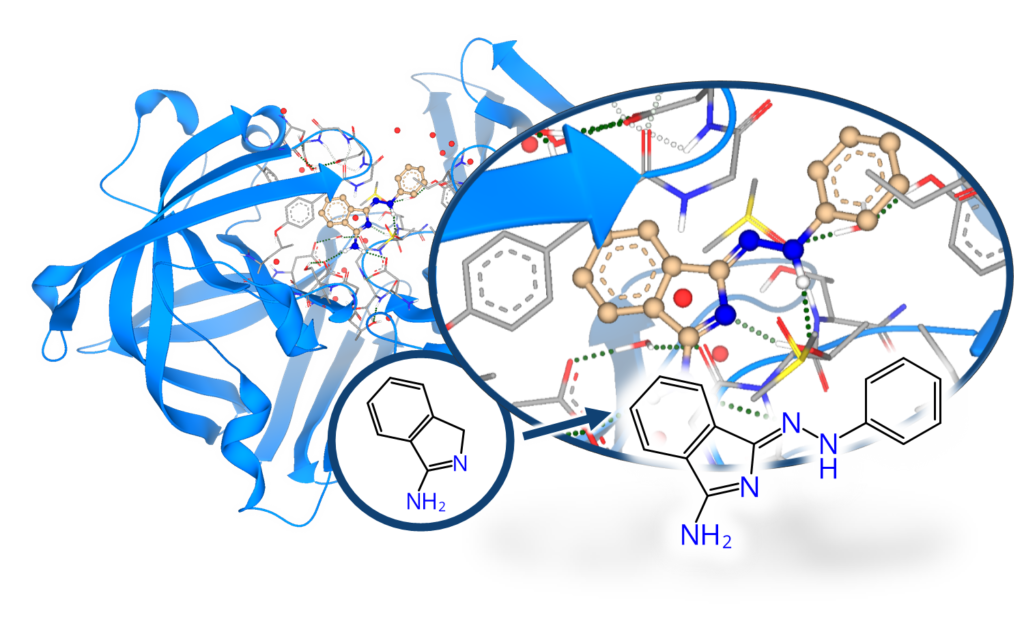
When fragment screening is carried out using X-ray crystallography it reveals the 3D-position of the fragment hits inside the protein’s binding site. This additional information of the fragment’s position is highly valuable for further improvement of the usual “low-affinity fragments” to create binders of higher potency. By combining fragment-based and structure-based drug discovery, binders of higher potency can be achieved.
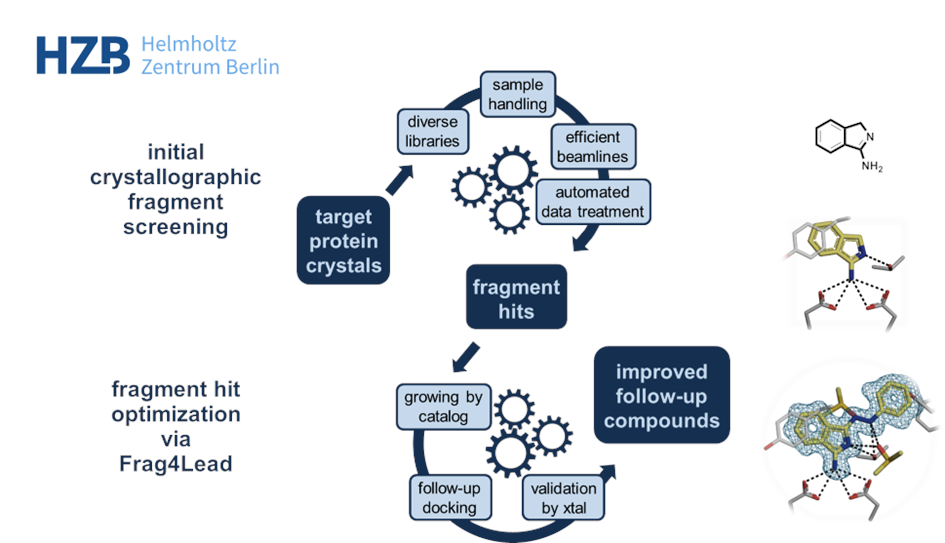
At the macromolecular crystallography (MX) beamlines at BESSY II, a dedicated workflow was established for the user community. It fosters efficient and convenient screening[1] and is based on several unique developments: First, the very diverse F2X fragment libraries that deliver high hit rates, mostly in the range of 20-25%.[2,3] Second, tools like the EasyAccess Frame ensure fast and comfortable crystal soaking and harvesting.[4] After data collection at the state-of-the-art MX beamlines at BESSY II, data analysis is highly automated and conveniently interfaced via the FragMAXapp setup at HZB.[5] FragMAXapp enables automatic data treatment using a number of pipelines, including the HZB-developments XDSAPP for automatic processing and fspipeline for automatic refinement.[6,7] As a final step, improved methodologies like PanDDA are applied for the best identification of the fragments in the electron density.[8]
Beyond efficient MX-based screening, HZB also offers methods of hit evolution to higher potency via fragment growing. In HZB’s Frag4Lead workflow the 3D-information of the crystallographic hits are used as an anchor for virtual pre-screening of suitable candidates from chemical catalogs.[9] This way, the first fragment growing step can be achieved without the need for custom synthesis and minimal virtual-screening expertise. Jan and team successfully employed Frag4Lead to advance fragment hits to single-digit micromolar binders in one step and shall report about this.
[1] Wollenhaupt, J. et al. J. Vis. Exp. 2021, 62208 (2021).
[2] Wollenhaupt, J. et al.. Structure 28, 694-706.e5 (2020).
[3] Barthel, T. et al. J. Med. Chem. (2022).
[4] Barthel, T. et al. J. Appl. Cryst. 54, 376–382 (2021).
[5] Lima, G. M. A. et al. Acta Cryst. D 77, 799–808 (2021).
[6] Sparta, K. et al. J. Appl. Cryst. 49, 1085-1092 (2016).
[7] Schiebel, J. et al. Structure 24, 1398–1409 (2016).
[8] Pearce, N. M., et al. Nature Comm. 8, 15123 (2017).
[9] Metz, A. et al. Acta Cryst. D 77, 1168–1182 (2021).